4JXM
 
 | | Crystal structure of RRP9 WD40 repeats | | Descriptor: | U3 small nucleolar RNA-interacting protein 2, UNKNOWN ATOM OR ION | | Authors: | Wu, X, Tempel, W, Xu, C, El Bakkouri, M, He, H, Seitova, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of RRP9 WD40 repeats
To be Published
|
|
2ONI
 
 | | Catalytic Domain of the Human NEDD4-like E3 Ligase | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like protein, SODIUM ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-24 | | Release date: | 2007-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NEDD4-like E3 Ubiquitin-protein Ligase
To be Published
|
|
1EJE
 
 | | CRYSTAL STRUCTURE OF AN FMN-BINDING PROTEIN | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, NICKEL (II) ION, ... | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Arrowsmith, C, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
3O4R
 
 | | Crystal Structure of Human Dehydrogenase/Reductase (SDR family) member 4 (DHRS4) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Dehydrogenase/reductase SDR family member 4, ... | | Authors: | Ugochukwu, E, Bhatia, C, Krojer, T, Vollmar, M, Yue, W.W, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Dehydrogenase/Reductase (SDR family) member 4 (DHRS4)
To be Published
|
|
1TU1
 
 | | Crystal Structure of Protein of Unknown Function PA94 from Pseudomonas aeruginosa, Putative Regulator | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-24 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of hypothetical protein PA94 from Pseudomonas aeruginosa
To be Published
|
|
1TUA
 
 | | 1.5 A Crystal Structure of a Protein of Unknown Function APE0754 from Aeropyrum pernix | | Descriptor: | Hypothetical protein APE0754 | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-24 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein APE0754 from Aeropyrum pernix
To be Published
|
|
1TP6
 
 | | 1.5 A Crystal Structure of a NTF-2 Like Protein of Unknown Function PA1314 from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein PA1314 | | Authors: | Zhang, R, Xu, L.X, savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-15 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA1314 from Pseudomonas aeruginosa
To be Published
|
|
3O46
 
 | | Crystal structure of the PDZ domain of MPP7 | | Descriptor: | MAGUK p55 subfamily member 7, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Tong, Y, Tempel, W, Zhong, N, Guan, X, Landry, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-26 | | Release date: | 2010-08-04 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the PDZ domain of MPP7
TO BE PUBLISHED
|
|
2OPG
 
 | | The crystal structure of the 10th PDZ domain of MPDZ | | Descriptor: | Multiple PDZ domain protein, SODIUM ION | | Authors: | Gileadi, C, Phillips, C, Elkins, J, Papagrigoriou, E, Ugochukwu, E, Gorrec, F, Savitsky, P, Umeano, C, Berridge, G, Gileadi, O, Salah, E, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the 10th PDZ domain of MPDZ
To be Published
|
|
1NE3
 
 | | Solution structure of ribosomal protein S28E from Methanobacterium Thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH0256_1_68; Northeast Structural Genomics Target TT744 | | Descriptor: | 30S ribosomal protein S28E | | Authors: | Wu, B, Pineda-Lucena, A, Yee, A, Cort, J.R, Ramelot, T.A, Kennedy, M, Edwards, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-10 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ribosomal protein S28E from Methanobacterium thermoautotrophicum.
Protein Sci., 12, 2003
|
|
1NC7
 
 | | Crystal Structure of Thermotoga maritima 1070 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Analysis of Thermotoga maritima Hypothetical protein TM1070
To be Published
|
|
2PBC
 
 | | FK506-binding protein 2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FK506-binding protein 2 | | Authors: | Walker, J.R, Neculai, D, Davis, T, Butler-Cole, C, Sicheri, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-28 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of FK506-Binding Protein 2
To be Published
|
|
2P02
 
 | | Crystal structure of the alpha subunit of human S-adenosylmethionine synthetase 2 | | Descriptor: | CHLORIDE ION, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthetase isoform type-2 | | Authors: | Papagrigoriou, E, Shafqat, N, Rojkova, A, Niessen, F.H, Kavanagh, K.L, von Delft, F, Gorrec, F, Ugochukwu, E, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal structure of the alpha subunit of human S-adenosylmethionine synthetase 2
To be Published
|
|
1H2H
 
 | | Crystal structure of TM1643 | | Descriptor: | HYPOTHETICAL PROTEIN TM1643, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yang, Z, Savchenko, A, Edwards, A, Arrowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-08-08 | | Release date: | 2002-08-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643.
J. Biol. Chem., 278, 2003
|
|
1P9Q
 
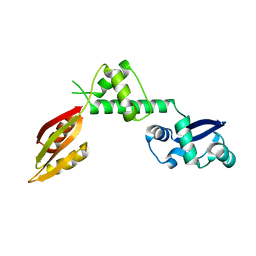 | | Structure of a hypothetical protein AF0491 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0491 | | Authors: | Savchenko, A, Evdokimova, E, Skarina, T, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A. | | Deposit date: | 2003-05-12 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism.
J.Biol.Chem., 280, 2005
|
|
3L43
 
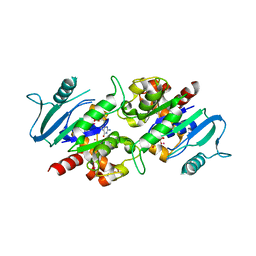 | | Crystal structure of the dynamin 3 GTPase domain bound with GDP | | Descriptor: | Dynamin-3, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Yang, S, Tempel, W, Tong, Y, Nedyalkova, L, Guan, X, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of the dynamin 3 GTPase domain bound with GDP
to be published
|
|
4IJD
 
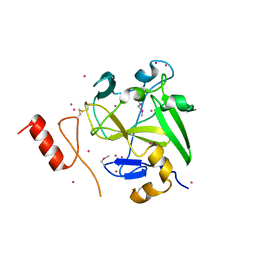 | | Crystal structure of methyltransferase domain of human PR domain-containing protein 9 | | Descriptor: | Histone-lysine N-methyltransferase PRDM9, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Dombrovski, L, Li, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-21 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of methyltransferase domain of human PR domain-containing protein 9
To be Published
|
|
1MKZ
 
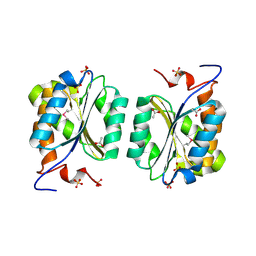 | | Crystal structure of MoaB protein at 1.6 A resolution. | | Descriptor: | ACETIC ACID, Molybdenum cofactor biosynthesis protein B, SULFATE ION | | Authors: | Sanishvili, R, Skarina, T, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Escherichia coli MoaB suggests a probable role in molybdenum cofactor synthesis.
J.Biol.Chem., 279, 2004
|
|
1M94
 
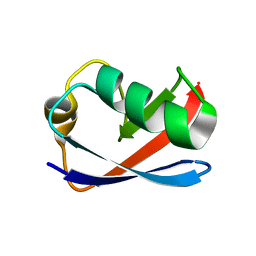 | | Solution Structure of the Yeast Ubiquitin-Like Modifier Protein Hub1 | | Descriptor: | Protein YNR032c-a | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Semesi, A, Edwards, A.M, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-07-26 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Yeast Ubiquitin-Like Modifier Protein Hub1
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
6ROG
 
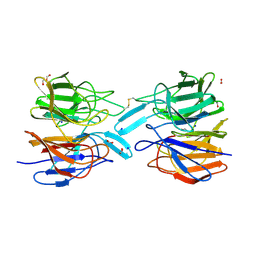 | | Crystal Structure of the KELCH domain of human KEAP1 | | Descriptor: | FORMIC ACID, Kelch-like ECH-associated protein 1, SODIUM ION | | Authors: | Sethi, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bullock, A.N, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structure of the KELCH domain of human KEAP1
To Be Published
|
|
3L5K
 
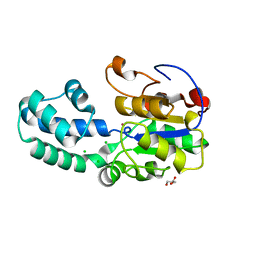 | | The crystal structure of human Haloacid Dehalogenase-like Hydrolase Domain containing 1A (HDHD1A) | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloacid dehalogenase-like hydrolase domain-containing protein 1A, ... | | Authors: | Ugochukwu, E, Guo, K, Picaud, S, Muniz, J, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Yue, W.W, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-22 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human Haloacid Dehalogenase-like Hydrolase Domain containing 1A (HDHD1A)
To be Published
|
|
3HE1
 
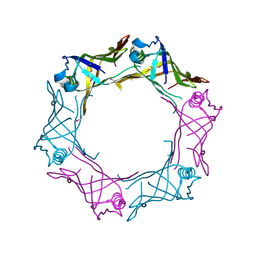 | | Secreted protein Hcp3 from Pseudomonas aeruginosa. | | Descriptor: | GLYCEROL, Major exported Hcp3 protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa.
J.Struct.Funct.Genom., 12, 2011
|
|
5OWR
 
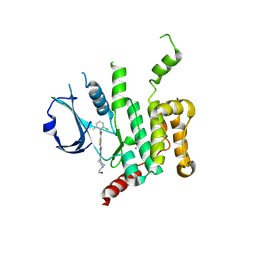 | | Human STK10 bound to dasatinib | | Descriptor: | CALCIUM ION, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, Serine/threonine-protein kinase 10 | | Authors: | Szklarz, M, Muniz, J.R.C, Vollmar, M, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2017-09-04 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human STK10 bound to dasatinib
To Be Published
|
|
1SH8
 
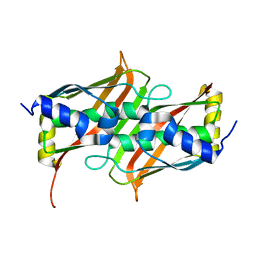 | | 1.5 A Crystal Structure of a Protein of Unknown Function PA5026 from Pseudomonas aeruginosa, Probable Thioesterase | | Descriptor: | hypothetical protein PA5026 | | Authors: | Zhang, R, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA5026 from Pseudomonas aeruginosa
To be Published
|
|
1RYK
 
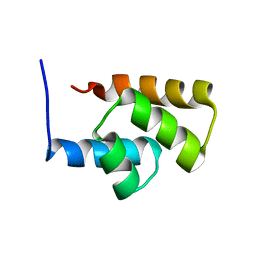 | | Solution NMR Structure Protein yjbJ from Escherichia coli. Northeast Structural Genomics Consortium Target ET93; Ontario Centre for Structural Proteomics target EC0298_1_69; | | Descriptor: | Protein yjbJ | | Authors: | Pineda-Lucena, A, Liao, J, Wu, B, Yee, A, Cort, J.R, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proteins, 47, 2002
|
|
