6M7K
 
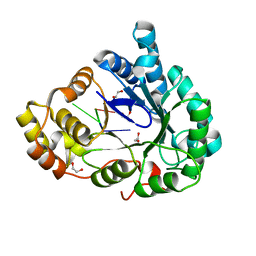 | | Structure of mouse RECON (AKR1C13) in complex with cyclic AMP-AMP-GMP (cAAG) | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C13, cyclic AMP-AMP-GMP | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-08-20 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
6EA9
 
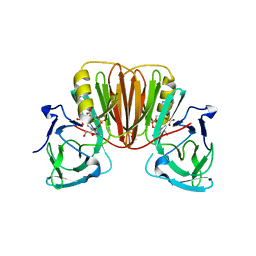 | |
6EA8
 
 | | Structure of VACV poxin in pre-reactive state with nonhydrolyzable 2'3' cGAMP | | Descriptor: | (2S,5R,7R,8R,10R,12aR,14R,15R,15aS,16R)-7-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-15,16-dihydroxy-2,10-disulfanyloctahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, O-[(1R,2R,3R)-5-{[(S)-{[(2R,3R,4R,5R)-2-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-4-hydroxy-5-(hydroxymethyl)tetrahydro furan-3-yl]oxy}(sulfanyl)phosphoryl]oxy}-1-(6-amino-9H-purin-9-yl)-1,2-dihydroxypentan-3-yl] dihydrogen (R)-phosphorothioate, Protein B2 | | Authors: | Eaglesham, J.B, Kranzusch, P.J. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling.
Nature, 566, 2019
|
|
6EA6
 
 | |
6E0K
 
 | | Structure of Rhodothermus marinus CdnE c-UMP-AMP synthase | | Descriptor: | cGAS/DncV-like nucleotidyltransferase in E. coli homolog | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
6E0L
 
 | | Structure of Rhodothermus marinus CdnE c-UMP-AMP synthase with Apcpp and Upnpp | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
6E0O
 
 | | Structure of Elizabethkingia meningoseptica CdnE cyclic dinucleotide synthase with pppA[3'-5']pA | | Descriptor: | MAGNESIUM ION, RNA (5'-D(*(ATP))-R(P*A)-3'), cGAS/DncV-like nucleotidyltransferase in E. coli homolog | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
6E0M
 
 | | Structure of Elizabethkingia meningoseptica CdnE cyclic dinucleotide synthase | | Descriptor: | DIPHOSPHATE, cGAS/DncV-like nucleotidyltransferase in E. coli homolog | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
6E0N
 
 | | Structure of Elizabethkingia meningoseptica CdnE cyclic dinucleotide synthase with GTP and Apcpp | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
6XB4
 
 | |
6XB6
 
 | |
6XB5
 
 | |
6XB3
 
 | |
7LJL
 
 | | Structure of the Enterobacter cloacae CD-NTase CdnD in complex with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-AMP-GMP synthase, ... | | Authors: | Govande, A, Lowey, B, Eaglesham, J.B, Whiteley, A.W, Kranzusch, P.J. | | Deposit date: | 2021-01-29 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis of CD-NTase nucleotide selection in CBASS anti-phage defense.
Cell Rep, 35, 2021
|
|
7LJM
 
 | | Structure of the Salmonella enterica CD-NTase CdnD in complex with GTP | | Descriptor: | CD-NTase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Govande, A, Lowey, B, Eaglesham, J.B, Whiteley, A.W, Kranzusch, P.J. | | Deposit date: | 2021-01-29 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of CD-NTase nucleotide selection in CBASS anti-phage defense.
Cell Rep, 35, 2021
|
|
7LJO
 
 | | Structure of the Bacteroides fragilis CD-NTase CdnB in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CD-NTase, MAGNESIUM ION | | Authors: | Govande, A, Lowey, B, Eaglesham, J.B, Whiteley, A.T, Kranzusch, P.J. | | Deposit date: | 2021-01-29 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis of CD-NTase nucleotide selection in CBASS anti-phage defense.
Cell Rep, 35, 2021
|
|
7LJN
 
 | | Structure of the Bradyrhizobium diazoefficiens CD-NTase CdnG in complex with GTP | | Descriptor: | CD-NTase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Govande, A, Lowey, B, Eaglesham, J.B, Whiteley, A.T, Kranzusch, P.J. | | Deposit date: | 2021-01-29 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of CD-NTase nucleotide selection in CBASS anti-phage defense.
Cell Rep, 35, 2021
|
|
