6HV5
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with 4 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6HW7
 
 | | Yeast 20S proteasome in complex with 29 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
5CZ9
 
 | |
6HVR
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with 16 | | Descriptor: | (2~{S})-~{N}-[(2~{S},3~{R})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-butan-2-yl]-2-[[(2~{R})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
5CT5
 
 | | Wild-type Bacillus subtilis lipase A with 10% [BMIM][Cl] | | Descriptor: | 1-butyl-3-methyl-1H-imidazol-3-ium, CHLORIDE ION, SULFATE ION, ... | | Authors: | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
6HWC
 
 | | Yeast 20S proteasome beta2-G45A mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6HVY
 
 | | Yeast 20S proteasome in complex with 5 (7- and 6-membered ring) | | Descriptor: | (2~{S})-~{N}-[(2~{S},3~{R})-1-[(1~{S},4~{a}~{S},8~{a}~{R})-1,2,3,4,4~{a},5,6,7,8,8~{a}-decahydronaphthalen-1-yl]-4-methyl-3,4-bis(oxidanyl)pentan-2-yl]-3-(4-methoxyphenyl)-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, (2~{S})-~{N}-[(2~{S},3~{S},4~{R})-1-[(1~{S},4~{a}~{S},8~{a}~{R})-1,2,3,4,4~{a},5,6,7,8,8~{a}-decahydronaphthalen-1-yl]-4-methyl-3,5-bis(oxidanyl)pentan-2-yl]-3-(4-methoxyphenyl)-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
5D61
 
 | | MOA-Z-VAD-fmk complex, direct orientation | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin, CALCIUM ION, ... | | Authors: | Cordara, G, van Eerde, A, Grahn, E.M, Goldstein, I.J, Krengel, U. | | Deposit date: | 2015-08-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Unusual Member of the Papain Superfamily: Mapping the Catalytic Cleft of the Marasmius oreades agglutinin (MOA) with a Caspase Inhibitor.
Plos One, 11, 2016
|
|
5BXN
 
 | | Yeast 20S proteasome beta2-G170A mutant in complex with Bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-06-09 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defective immuno- and thymoproteasome assembly causes severe immunodeficiency.
Sci Rep, 8, 2018
|
|
7BNT
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikD with a predicted ancestral HMA domain of Pik-1 from Oryza spp. | | Descriptor: | 1,2-ETHANEDIOL, AVR-Pik protein, Predicted ancestral HMA domain of Pik-1 from Oryza spp. | | Authors: | Bialas, A, Langner, T, Harant, A, Contreras, M.P, Stevenson, C.E.M, Lawson, D.M, Sklenar, J, Kellner, R, Moscou, M.J, Terauchi, R, Banfield, M.J, Kamoun, S. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Two NLR immune receptors acquired high-affinity binding to a fungal effector through convergent evolution of their integrated domain.
Elife, 10, 2021
|
|
5FBH
 
 | | Crystal structure of the extracellular domain of human calcium sensing receptor with bound Gd3+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Zhang, T, Zhang, C, Miller, C.L, Zou, J, Moremen, K.W, Brown, E.M, Yang, J.J, Hu, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist.
Sci Adv, 2, 2016
|
|
5FTY
 
 | | Structure of surface layer protein SbsC, domains 6-7 (monoclinic form) | | Descriptor: | CALCIUM ION, SURFACE LAYER PROTEIN | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2016-01-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Surface Layer Protein Sbsc, Domains 6-7 (Monoclinic Form)
To be Published
|
|
5FGE
 
 | |
5FGA
 
 | |
5FG7
 
 | | Yeast 20S proteasome beta2-T1A mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-12-20 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
5FG9
 
 | | Yeast 20S proteasome beta2-T(-2)V mutant | | Descriptor: | MAGNESIUM ION, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-12-20 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
5FHS
 
 | |
5FGF
 
 | |
5FBK
 
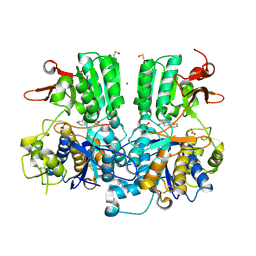 | | Crystal structure of the extracellular domain of human calcium sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Zhang, T, Zhang, C, Miller, C.L, Zou, J, Moremen, K.W, Brown, E.M, Yang, J.J, Hu, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist.
Sci Adv, 2, 2016
|
|
3JD5
 
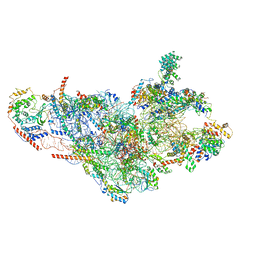 | | Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome | | Descriptor: | 28S ribosomal RNA, mitochondial, 28S ribosomal protein S10, ... | | Authors: | Kaushal, P.S, Sharma, M.R, Booth, T.M, Haque, E.M, Tung, C.S, Sanbonmatsu, K.Y, Spremulli, L.L, Agrawal, R.K. | | Deposit date: | 2016-04-08 | | Release date: | 2016-07-13 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7T6Y
 
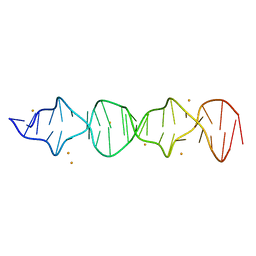 | | d((CGA)5TGA) parallel-stranded homo-duplex | | Descriptor: | BARIUM ION, DNA (5'-D(*CP*GP*AP*CP*GP*AP*CP*GP*AP*CP*GP*AP*CP*GP*AP*TP*GP*A)-3') | | Authors: | Luteran, E.M, Paukstelis, P.J. | | Deposit date: | 2021-12-14 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The parallel-stranded d(CGA) duplex is a highly predictable structural motif with two conformationally distinct strands.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7T4Q
 
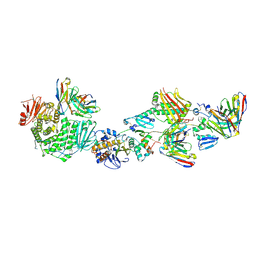 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with neutralizing fabs 2C12, 7I13 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4S
 
 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with NRP2 and neutralizing fabs 8I21 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4R
 
 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with THBD and neutralizing fabs MSL-109 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
4Q9C
 
 | | IgNAR antibody domain C3 | | Descriptor: | CHLORIDE ION, Novel antigen receptor, SODIUM ION, ... | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
