1ZDQ
 
 | | Crystal Structure of Met150Gly AfNiR with Methylsulfanyl Methane Bound | | Descriptor: | (METHYLSULFANYL)METHANE, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Wijma, H.J, MacPherson, I.S, Alexandre, M, Diederix, R.E.M, Canters, G.W, Murphy, M.E.P, Verbeet, M.P. | | Deposit date: | 2005-04-14 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A rearranging ligand enables allosteric control of catalytic activity in copper-containing nitrite reductase.
J.Mol.Biol., 358, 2006
|
|
1ZFJ
 
 | | INOSINE MONOPHOSPHATE DEHYDROGENASE (IMPDH; EC 1.1.1.205) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | INOSINE MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID | | Authors: | Zhang, R, Evans, G, Rotella, F.J, Westbrook, E.M, Beno, D, Huberman, E, Joachimiak, A, Collart, F.R. | | Deposit date: | 1999-03-29 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characteristics and crystal structure of bacterial inosine-5'-monophosphate dehydrogenase.
Biochemistry, 38, 1999
|
|
1YWA
 
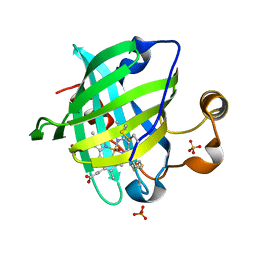 | | 0.9 A Structure of NP4 from Rhodnius Prolixus complexed with CO at pH 5.6 | | Descriptor: | CARBON MONOXIDE, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Maes, E.M, Weichsel, A, Roberts, S.A, Montfort, W.R. | | Deposit date: | 2005-02-17 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Ultrahigh Resolution Structures of Nitrophorin 4: Heme Distortion in Ferrous CO and NO Complexes
Biochemistry, 44, 2005
|
|
1ZY8
 
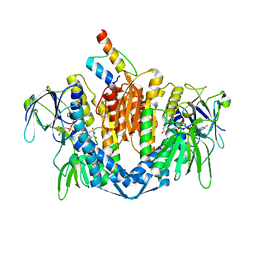 | | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ciszak, E.M, Makal, A, Hong, Y.S, Vettaikkorumakankauv, A.K, Korotchkina, L.G, Patel, M.S. | | Deposit date: | 2005-06-09 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | How Dihydrolipoamide Dehydrogenase-binding Protein Binds Dihydrolipoamide Dehydrogenase in the Human Pyruvate Dehydrogenase Complex.
J.Biol.Chem., 281, 2006
|
|
6HV4
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with ONX 0914 | | Descriptor: | (2~{S})-3-(4-methoxyphenyl)-~{N}-[(2~{S},3~{R})-4-methyl-3,4-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6HW4
 
 | | Yeast 20S proteasome in complex with 16 | | Descriptor: | (2~{S})-~{N}-[(2~{S},3~{R})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-butan-2-yl]-2-[[(2~{R})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6HVA
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with 13 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6HW9
 
 | | Yeast 20S proteasome in complex with 41b | | Descriptor: | (2~{S})-3-(4-methoxyphenyl)-~{N}-[(2~{S},3~{R})-4-methyl-1-(4-methylcyclohexyl)-3,4-bis(oxidanyl)pentan-2-yl]-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
1N66
 
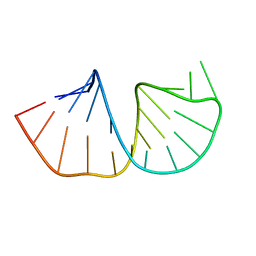 | | Structure of the pyrimidine-rich internal loop in the Y-domain of poliovirus 3'UTR | | Descriptor: | internal loop in the Y-domain of poliovirus 3'UTR | | Authors: | Lescrinier, E.M, Tessari, M, van Kuppeveld, F.J, Melchers, W.J, Hilbers, C.W, Heus, H.A. | | Deposit date: | 2002-11-08 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Pyrimidine-rich Internal Loop in the Poliovirus 3'-UTR: The Importance of Maintaining Pseudo-2-fold Symmetry in RNA Helices Containing Two Adjacent Non-canonical Base-pairs.
J.Mol.Biol., 331, 2003
|
|
6HW3
 
 | | Yeast 20S proteasome in complex with 13 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
1XOW
 
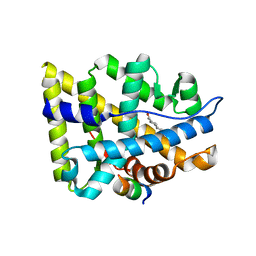 | | Crystal structure of the human androgen receptor ligand binding domain bound with an androgen receptor NH2-terminal peptide, AR20-30, and R1881 | | Descriptor: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, androgen receptor, decamer fragment of androgen receptor | | Authors: | He, B, Gampe Jr, R.T, Kole, A.J, Hnat, A.T, Stanley, T.B, An, G, Stewart, E.L, Kalman, R.I, Minges, J.T, Wilson, E.M. | | Deposit date: | 2004-10-07 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance
Mol.Cell, 16, 2004
|
|
2Q20
 
 | |
1OGV
 
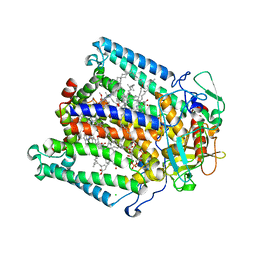 | | Lipidic cubic phase crystal structure of the photosynthetic reaction centre from Rhodobacter sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Katona, G, Andreasson, U, Landau, E.M, Andreasson, L.-E, Neutze, R. | | Deposit date: | 2003-05-13 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Lipidic Cubic Phase Crystal Structure of the Photosynthetic Reaction Centre from Rhodobacter Sphaeroides at 2.35 A Resolution
J.Mol.Biol., 331, 2003
|
|
1OGU
 
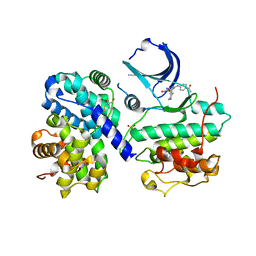 | | STRUCTURE OF HUMAN THR160-PHOSPHO CDK2/CYCLIN A COMPLEXED WITH A 2-ARYLAMINO-4-CYCLOHEXYLMETHYL-5-NITROSO-6-AMINOPYRIMIDINE INHIBITOR | | Descriptor: | 4-{[4-AMINO-6-(CYCLOHEXYLMETHOXY)-5-NITROSOPYRIMIDIN-2-YL]AMINO}BENZAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-05-13 | | Release date: | 2003-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of 2-Arylamino-4-Cyclohexyl Methyl-5-Nitroso-6-Aminopyrimidine Inhibitors of Cyclin-Dependent Kinases 1 and 2
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1YFH
 
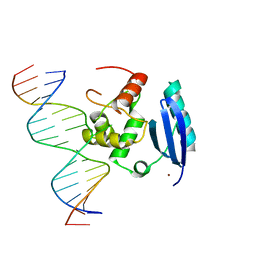 | | wt Human O6-Alkylguanine-DNA Alkyltransferase Bound To DNA Containing an Alkylated Cytosine | | Descriptor: | 5'-D(*CP*CP*TP*AP*CP*AP*CP*AP*CP*AP*TP*CP*CP*AP*CP*A)-3', 5'-D(*GP*TP*GP*GP*AP*TP*GP*(XCY)P*GP*TP*GP*TP*AP*GP*GP*T)-3', Methylated-DNA--protein-cysteine methyltransferase, ... | | Authors: | Duguid, E.M, Rice, P.A, He, C. | | Deposit date: | 2004-12-31 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | The structure of the human AGT protein bound to DNA and its implications for damage detection.
J.Mol.Biol., 350, 2005
|
|
1YH2
 
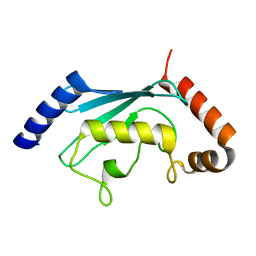 | | Ubiquitin-Conjugating Enzyme HSPC150 | | Descriptor: | HSPC150 protein similar to ubiquitin-conjugating enzyme | | Authors: | Walker, J.R, Avvakumov, G.V, Newman, E.M, Mackenzie, F, Kozieradzki, I, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
1QHJ
 
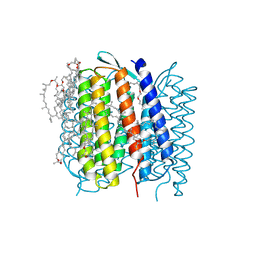 | | X-RAY STRUCTURE OF BACTERIORHODOPSIN GROWN IN LIPIDIC CUBIC PHASES | | Descriptor: | 1,2-[DI-2,6,10,14-TETRAMETHYL-HEXADECAN-16-OXY]-PROPANE, PROTEIN (BACTERIORHODOPSIN), RETINAL | | Authors: | Belrhali, H, Nollert, P, Royant, A, Menzel, C, Rosenbusch, J.P, Landau, E.M, Pebay-Peyroula, E. | | Deposit date: | 1999-05-04 | | Release date: | 1999-07-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein, lipid and water organization in bacteriorhodopsin crystals: a molecular view of the purple membrane at 1.9 A resolution.
Structure Fold.Des., 7, 1999
|
|
1ZUO
 
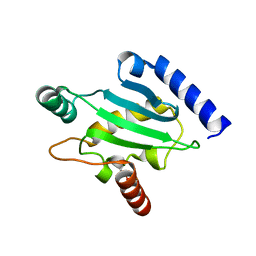 | | Structure of Human Ubiquitin-Conjugating Enzyme (UBCi) Involved in Embryo Attachment and Implantation | | Descriptor: | BETA-MERCAPTOETHANOL, Hypothetical protein LOC92912 | | Authors: | Walker, J.R, Avvakumov, G.V, Cui, H, Newman, E.M, Mackenzie, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-31 | | Release date: | 2005-07-05 | | Last modified: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
2PHK
 
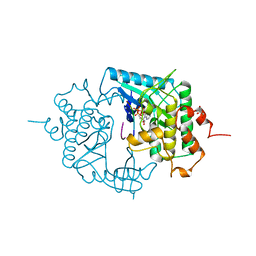 | | THE CRYSTAL STRUCTURE OF A PHOSPHORYLASE KINASE PEPTIDE SUBSTRATE COMPLEX: KINASE SUBSTRATE RECOGNITION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lowe, E.D, Noble, M.E.M, Skamnaki, V.T, Oikonomakos, N.G, Owen, D.J, Johnson, L.N. | | Deposit date: | 1998-06-18 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition.
EMBO J., 16, 1997
|
|
2A7M
 
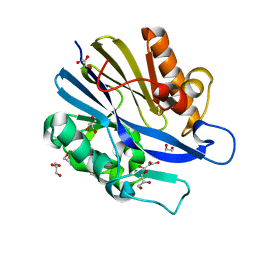 | | 1.6 Angstrom Resolution Structure of the Quorum-Quenching N-Acyl Homoserine Lactone Hydrolase of Bacillus thuringiensis | | Descriptor: | GLYCEROL, N-acyl homoserine lactone hydrolase, ZINC ION | | Authors: | Liu, D, Lepore, B.W, Petsko, G.A, Thomas, P.W, Stone, E.M, Fast, W, Ringe, D. | | Deposit date: | 2005-07-05 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-dimensional structure of the quorum-quenching N-acyl homoserine lactone hydrolase from Bacillus thuringiensis
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1OW8
 
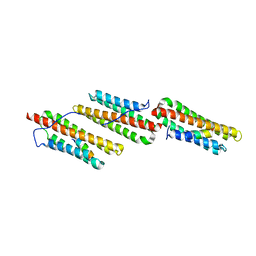 | | Paxillin LD2 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular Recognition of Paxillin LD Motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
2A4O
 
 | | Dual modes of modification of Hepatitis A virus 3C protease by a serine derived beta-lactone: selective crytstallization and high resolution structure of the His102 adduct | | Descriptor: | ACETYL GROUP, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, PHENYLALANINE AMIDE, ... | | Authors: | Yin, J, Bergmann, E.M, Cherney, M.M, Lall, M.S, Jain, R.P, Vederas, J.C, James, M.N.G. | | Deposit date: | 2005-06-29 | | Release date: | 2005-12-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-derived beta-Lactone: Selective Crystallization and Formation of a Functional Catalytic Triad in the Active Site
J.MOL.BIOL., 354, 2005
|
|
6HQ8
 
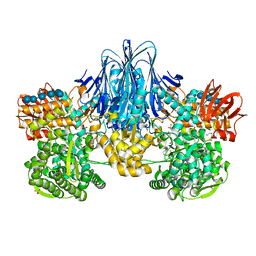 | | Bacterial beta-1,3-oligosaccharide phosphorylase from GH149 with laminarihexaose bound at a surface site | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Beta-1,3-oligosaccharide phosphorylase, ... | | Authors: | Kuhaudomlarp, S, Stevenson, C.E.M, Lawson, D.M, Field, R.A. | | Deposit date: | 2018-09-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of a GH149 beta-(1 → 3) glucan phosphorylase reveals a new surface oligosaccharide binding site and additional domains that are absent in the disaccharide-specific GH94 glucose-beta-(1 → 3)-glucose (laminaribiose) phosphorylase.
Proteins, 87, 2019
|
|
6HTD
 
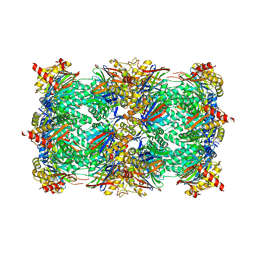 | | Yeast 20S proteasome with human beta2c (S171G) in complex with 4 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-03 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6HUU
 
 | |
