3OJE
 
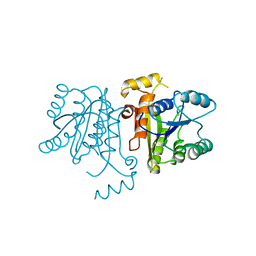 | | Crystal Structure of the Bacillus cereus Enoyl-Acyl Carrier Protein Reductase (Apo form) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase (FabL) (NADPH) | | Authors: | Kim, S.J, Ha, B.H, Kim, K.H, Hong, S.K, Suh, S.W, Kim, E.E. | | Deposit date: | 2010-08-22 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Dimeric and tetrameric forms of enoyl-acyl carrier protein reductase from Bacillus cereus
Biochem.Biophys.Res.Commun., 400, 2010
|
|
3OZ3
 
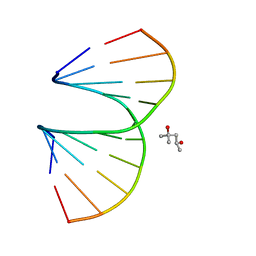 | | Vinyl Carbocyclic LNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*(UVX)P*AP*CP*GP*C)-3') | | Authors: | Seth, P.R, Allerson, C.A, Berdeja, A, Siwkowski, A, Pallan, P.S, Gaus, H, Prakash, T.P, Watt, A.T, Egli, M, Swayze, E.E. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA.
J.Am.Chem.Soc., 132, 2010
|
|
3OQC
 
 | |
3OZ5
 
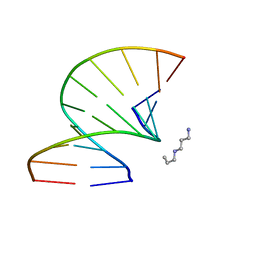 | | S-Methyl Carbocyclic LNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(UMX)P*AP*CP*GP*C)-3'), SPERMINE | | Authors: | Seth, P.R, Allerson, C.A, Berdeja, A, Siwkowski, A, Pallan, P.S, Gaus, H, Prakash, T.P, Watt, A.T, Egli, M, Swayze, E.E. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA.
J.Am.Chem.Soc., 132, 2010
|
|
3OID
 
 | | Crystal Structure of Enoyl-ACP Reductases III (FabL) from B. subtilis (complex with NADP and TCL) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRICLOSAN | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3OIG
 
 | | Crystal Structure of Enoyl-ACP Reductases I (FabI) from B. subtilis (complex with NAD and INH) | | Descriptor: | (2E)-N-[(1,2-dimethyl-1H-indol-3-yl)methyl]-N-methyl-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3OJF
 
 | | Crystal Structure of the Bacillus cereus Enoyl-Acyl Carrier Protein Reductase with NADP+ and indole naphthyridinone (Complex form) | | Descriptor: | (2E)-N-[(1,2-dimethyl-1H-indol-3-yl)methyl]-N-methyl-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase (FabL) (NADPH), NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kim, S.J, Ha, B.H, Kim, K.H, Hong, S.K, Suh, S.W, Kim, E.E. | | Deposit date: | 2010-08-22 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dimeric and tetrameric forms of enoyl-acyl carrier protein reductase from Bacillus cereus
Biochem.Biophys.Res.Commun., 400, 2010
|
|
3OZ4
 
 | | R-Methyl Carbocyclic LNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(URX)P*AP*CP*GP*C)-3') | | Authors: | Seth, P.R, Allerson, C.A, Berdeja, A, Siwkowski, A, Pallan, P.S, Gaus, H, Prakash, T.P, Watt, A.T, Egli, M, Swayze, E.E. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA.
J.Am.Chem.Soc., 132, 2010
|
|
3FW0
 
 | | Structure of Peptidyl-alpha-hydroxyglycine alpha-Amidating Lyase (PAL) bound to alpha-hydroxyhippuric acid (non-peptidic substrate) | | Descriptor: | (2S)-hydroxy[(phenylcarbonyl)amino]ethanoic acid, CALCIUM ION, MERCURY (II) ION, ... | | Authors: | Chufan, E.E, De, M, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2009-01-16 | | Release date: | 2009-09-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme.
Structure, 17, 2009
|
|
3FVZ
 
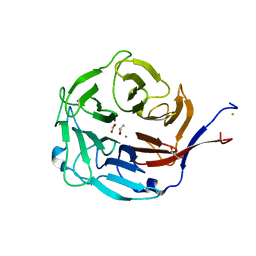 | | Structure of Peptidyl-alpha-hydroxyglycine alpha-Amidating Lyase (PAL) | | Descriptor: | ACETATE ION, CALCIUM ION, FE (III) ION, ... | | Authors: | Chufan, E.E, De, M, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2009-01-16 | | Release date: | 2009-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme.
Structure, 17, 2009
|
|
3OIF
 
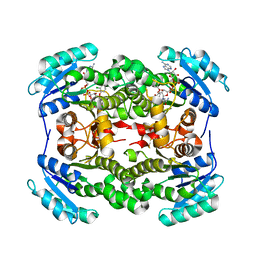 | | Crystal Structure of Enoyl-ACP Reductases I (FabI) from B. subtilis (complex with NAD and TCL) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3QX1
 
 | | Crystal structure of FAF1 UBX domain | | Descriptor: | FAS-associated factor 1, SULFATE ION | | Authors: | Park, J.K, Jeon, H, Lee, J.J, Kim, K.H, Lee, K.J, Kim, E.E. | | Deposit date: | 2011-03-01 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dissection of the interaction between FAF1 UBX and p97
To be Published
|
|
3QOQ
 
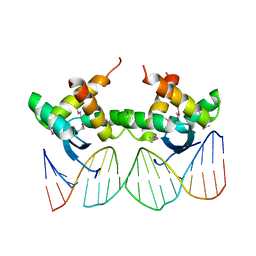 | |
3QWZ
 
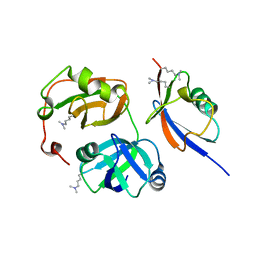 | | Crystal structure of FAF1 UBX-p97N-domain complex | | Descriptor: | FAS-associated factor 1, Transitional endoplasmic reticulum ATPase | | Authors: | Park, J.K, Jeon, H, Lee, J.J, Kim, K.H, Lee, K.J, Kim, E.E. | | Deposit date: | 2011-02-28 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissection of the interaction between FAF1 UBX and p97
To be Published
|
|
3Q61
 
 | | 3'-Fluoro Hexitol Nucleic Acid DNA Structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(F3H)P*AP*CP*GP*C)-3') | | Authors: | Seth, P.R, Allerson, C.R, Prakash, T.P, Siwkowski, A, Berdeja, A, Yu, J, Pallan, P.S, Watt, A.T, Gaus, H, Bhat, B, Egli, M, Swayze, E.E. | | Deposit date: | 2010-12-30 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Synthesis, improved antisense activity and structural rationale for the divergent RNA affinities of 3'-fluoro hexitol nucleic acid (FHNA and Ara-FHNA) modified oligonucleotides.
J.Am.Chem.Soc., 133, 2011
|
|
3IM9
 
 | | Crystal structure of MCAT from Staphylococcus aureus | | Descriptor: | ACETATE ION, CALCIUM ION, Malonyl CoA-acyl carrier protein transacylase, ... | | Authors: | Hong, S.K, Kim, K.H, Park, J.K, Kim, Y.M, Kim, E.E. | | Deposit date: | 2009-08-10 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | New design platform for malonyl-CoA-acyl carrier protein transacylase
Febs Lett., 584, 2010
|
|
3IM8
 
 | | Crystal structure of MCAT from Streptococcus pneumoniae | | Descriptor: | ACETATE ION, Malonyl acyl carrier protein transacylase | | Authors: | Hong, S.K, Kim, K.H, Park, J.K, Kim, Y.M, Kim, E.E. | | Deposit date: | 2009-08-10 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New design platform for malonyl-CoA-acyl carrier protein transacylase
Febs Lett., 584, 2010
|
|
3ITQ
 
 | | Crystal Structure of a Prolyl 4-Hydroxylase from Bacillus anthracis | | Descriptor: | GLYCEROL, PHOSPHATE ION, Prolyl 4-hydroxylase, ... | | Authors: | Culpepper, M.A, Scott, E.E, Limburg, J. | | Deposit date: | 2009-08-28 | | Release date: | 2009-12-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of prolyl 4-hydroxylase from Bacillus anthracis.
Biochemistry, 49, 2010
|
|
3IQB
 
 | | Tt I75F/L144F H-NOX | | Descriptor: | Methyl-accepting chemotaxis protein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Weinert, E.E, Plate, L, Whited, C.A, Olea Jr, C, Marletta, M.A. | | Deposit date: | 2009-08-19 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Determinants of Ligand Affinity and Heme Reactivity in H-NOX Domains.
Angew.Chem.Int.Ed.Engl., 49, 2009
|
|
3RBY
 
 | |
3J05
 
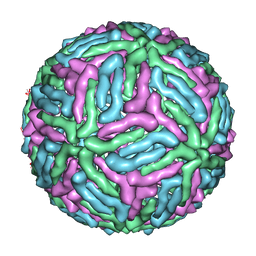 | | Three-dimensional structure of Dengue virus serotype 1 complexed with HMAb 14c10 Fab | | Descriptor: | envelope protein | | Authors: | Teoh, E.P, Kukkaro, P, Teo, E.W, Lim, A, Tan, T.T, Shi, P.Y, Yip, A, Schul, W, Leo, Y.S, Chan, S.H, Smith, K.G.C, Ooi, E.E, Kemeny, D.M, Ng, G, Ng, M.L, Alonso, S, Fisher, D, Hanson, B, Lok, S.M, MacAry, P.A. | | Deposit date: | 2011-04-01 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | The structural basis for serotype-specific neutralization of dengue virus by a human antibody.
Sci Transl Med, 4, 2012
|
|
3RUK
 
 | | Human Cytochrome P450 CYP17A1 in complex with Abiraterone | | Descriptor: | Abiraterone, PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of cytochrome P450 17A1 with prostate cancer drugs abiraterone and TOK-001.
Nature, 482, 2012
|
|
3KDB
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10006 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Chufan, E.E, Lafont, V, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
3KDC
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10074 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dichlorophenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chufan, E.E, Kawasaki, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
3KDD
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10265 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-difluorophenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H- inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Chufan, E.E, Kawasaki, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
