5WTE
 
 | | Cryo-EM structure for Hepatitis A virus full particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTG
 
 | | Crystal structure of the Fab fragment of anti-HAV antibody R10 | | Descriptor: | FAB Heavy chain, FAB Light chain | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5X3S
 
 | |
5B6N
 
 | |
5MQW
 
 | | High-speed fixed-target serial virus crystallography | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Roedig, P, Ginn, H.M, Pakendorf, T, Sutton, G, Harlos, K, Walter, T.S, Meyer, J, Fischer, P, Duman, R, Vartiainen, I, Reime, B, Warmer, M, Brewster, A.S, Young, I.D, Michels-Clark, T, Sauter, N.K, Sikorsky, M, Nelson, S, Damiani, D.S, Alonso-Mori, R, Ren, J, Fry, E.E, David, C, Stuart, D.I, Wagner, A, Meents, A. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-speed fixed-target serial virus crystallography.
Nat. Methods, 14, 2017
|
|
6WR1
 
 | | Human steroidogenic cytochrome P450 17A1 mutant N52Y with inhibitor abiraterone | | Descriptor: | Abiraterone, PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase | | Authors: | Petrunak, E.M, Bart, A.G, Scott, E.E. | | Deposit date: | 2020-04-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Human cytochrome P450 17A1 structures with metabolites of prostate cancer drug abiraterone reveal substrate-binding plasticity and a second binding site.
J.Biol.Chem., 299, 2023
|
|
6WR0
 
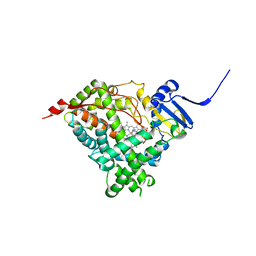 | | Human steroidogenic cytochrome P450 17A1 with 3-keto-delta4-abiraterone analog | | Descriptor: | (8alpha)-17-(pyridin-3-yl)androsta-4,16-dien-3-one, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Petrunak, E.M, Bart, A.G, Scott, E.E. | | Deposit date: | 2020-04-29 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human cytochrome P450 17A1 structures with metabolites of prostate cancer drug abiraterone reveal substrate-binding plasticity and a second binding site.
J.Biol.Chem., 299, 2023
|
|
6WW0
 
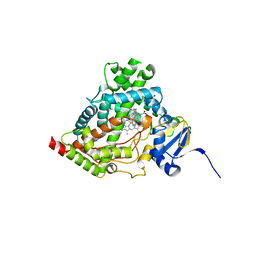 | | Human steroidogenic cytochrome P450 17A1 with 3-keto-5alpha-abiraterone analog | | Descriptor: | (5alpha,8alpha)-17-(pyridin-3-yl)androst-16-en-3-one, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Petrunak, E.M, Bart, A.G, Scott, E.E. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Human cytochrome P450 17A1 structures with metabolites of prostate cancer drug abiraterone reveal substrate-binding plasticity and a second binding site.
J.Biol.Chem., 299, 2023
|
|
5N1J
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Giastas, P, Andreou, A, Balomenou, S, Bouriotis, V, Eliopoulos, E.E. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5NFQ
 
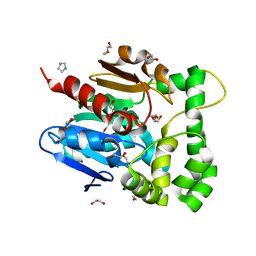 | | Novel epoxide hydrolases belonging to the alpha/beta hydrolases superfamily in metagenomes from hot environments | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Ferrandi, E.E, De Rose, S.A, Sayer, C, Guazzelli, E, Marchesi, C, Saneei, V, Isupov, M.N, Littlechild, J.A, Monti, D. | | Deposit date: | 2017-03-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Thermophilic alpha / beta Class Epoxide Hydrolases Found in Metagenomes From Hot Environments.
Front Bioeng Biotechnol, 6, 2018
|
|
5NEK
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, ACETATE ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Andreou, A, Giastas, P, Eliopoulos, E.E. | | Deposit date: | 2017-03-10 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.057 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
3T3S
 
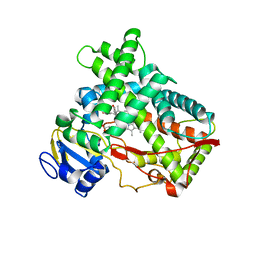 | | Human Cytochrome P450 2A13 in complex with Pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2A13, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
3T3R
 
 | | Human Cytochrome P450 2A6 in complex with Pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
6XYH
 
 | | NMR solution structure of alpha-AnmTX-Ms11a-2 (Ms11a-2) | | Descriptor: | AMS3 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Logashina, Y.A, Maleeva, E.E, Andreev, Y.A. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-AnmTX-Ms11a-2 (Ms11a-2)
To Be Published
|
|
5N1P
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with N-hydroxynaphthalene-1-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, Peptidoglycan N-acetylglucosamine deacetylase, SODIUM ION, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5NCD
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with (2S)-2-amino-5-(diaminomethylideneamino)-N-hydroxypentanamide | | Descriptor: | ACETATE ION, CITRIC ACID, N-HYDROXY-L-ARGININAMIDE, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5NG7
 
 | | Novel epoxide hydrolases belonging to the alpha/beta hydrolases superfamily in metagenomes from hot environments | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ferrandi, E.E, De Rose, S.A, Sayer, C, Guazzelli, E, Marchesi, C, Saneei, V, Isupov, M.N, Littlechild, J.A, Monti, D. | | Deposit date: | 2017-03-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Thermophilic alpha / beta Class Epoxide Hydrolases Found in Metagenomes From Hot Environments.
Front Bioeng Biotechnol, 6, 2018
|
|
5NC6
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with (E)-N-hydroxy-3-(naphthalen-1-yl)prop-2-enamide | | Descriptor: | 1,2-ETHANEDIOL, 3-naphthalen-1-yl-~{N}-oxidanyl-propanamide, ACETATE ION, ... | | Authors: | Giastas, P, Andreou, A, Balomenou, S, Bouriotis, V, Eliopoulos, E.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5NEL
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with ThiametG | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Andreou, A, Giastas, P, Eliopoulos, E.E. | | Deposit date: | 2017-03-10 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
6YM0
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab (crystal form 1) | | Descriptor: | Spike glycoprotein, heavy chain, light chain | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.36 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6YOR
 
 | | Structure of the SARS-CoV-2 spike S1 protein in complex with CR3022 Fab | | Descriptor: | IgG H chain, IgG L chain, Spike glycoprotein | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Duyvesteyn, H.M.E, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-15 | | Release date: | 2020-04-29 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
3SJ5
 
 | | I5F Mutant Structure of T. Tengcongensis H-NOX | | Descriptor: | Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weinert, E.E, Phillips-Piro, C.M, Tran, R, Mathies, R.A, Marletta, M.A. | | Deposit date: | 2011-06-20 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Controlling Conformational Flexibility of an O2-Binding H-NOX Domain.
Biochemistry, 50, 2011
|
|
6YLA
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
5NC9
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with (2S)-2,6-diamino-N-hydroxyhexanamide | | Descriptor: | (2~{S})-2,6-bis(azanyl)-~{N}-oxidanyl-hexanamide, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
6XYI
 
 | | NMR solution structure of alpha-AnmTX- Ms11a-3 (Ms11a-3) | | Descriptor: | AMS9.3.1 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Logashina, Y.A, Maleeva, E.E, Andreev, Y.A. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-AnmTX- Ms11a-3 (Ms11a-3)
TO BE PUBLISHED
|
|
