1RR9
 
 | | Catalytic domain of E.coli Lon protease | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2003-12-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1STA
 
 | | ACCOMMODATION OF INSERTION MUTATIONS ON THE SURFACE AND IN THE INTERIOR OF STAPHYLOCOCCAL NUCLEASE | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Keefe, L.J, Quirk, S, Gittis, A, Lattman, E.E. | | Deposit date: | 1994-01-17 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accommodation of insertion mutations on the surface and in the interior of staphylococcal nuclease.
Protein Sci., 3, 1994
|
|
1SZ3
 
 | | CRYSTAL STRUCTURE OF NUDIX HYDROLASE DR1025 IN COMPLEXED WITH GNP AND MG+2 | | Descriptor: | MAGNESIUM ION, MutT/nudix family protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-04-02 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
1RNU
 
 | | REFINEMENT OF THE CRYSTAL STRUCTURE OF RIBONUCLEASE S. COMPARISON WITH AND BETWEEN THE VARIOUS RIBONUCLEASE A STRUCTURES | | Descriptor: | RIBONUCLEASE S, SULFATE ION | | Authors: | Kim, E.E, Varadarajan, R, Wyckoff, H.W, Richards, F.M. | | Deposit date: | 1992-02-19 | | Release date: | 1994-01-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refinement of the crystal structure of ribonuclease S. Comparison with and between the various ribonuclease A structures.
Biochemistry, 31, 1992
|
|
1SU2
 
 | | CRYSTAL STRUCTURE OF THE NUDIX HYDROLASE DR1025 IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MutT/nudix family protein | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
1SUO
 
 | | Structure of mammalian cytochrome P450 2B4 with bound 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Scott, E.E, White, M.A, He, Y.A, Johnson, E.F, Stout, C.D, Halpert, J.R. | | Deposit date: | 2004-03-26 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9 {angstrom} resolution: Insight into the range of P450 conformations and coordination of redox partner binding.
J.Biol.Chem., 279, 2004
|
|
1TCO
 
 | | TERNARY COMPLEX OF A CALCINEURIN A FRAGMENT, CALCINEURIN B, FKBP12 AND THE IMMUNOSUPPRESSANT DRUG FK506 (TACROLIMUS) | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, FE (III) ION, ... | | Authors: | Griffith, J.P, Kim, J.L, Kim, E.E, Sintchak, M.D, Thomson, J.A, Fitzgibbon, M.J, Fleming, M.A, Caron, P.R, Hsiao, K, Navia, M.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex.
Cell(Cambridge,Mass.), 82, 1995
|
|
1STY
 
 | |
1STH
 
 | | TWO DISTINCTLY DIFFERENT METAL BINDING MODES ARE SEEN IN X-RAY CRYSTAL STRUCTURES OF STAPHYLOCOCCAL NUCLEASE-COBALT(II)-NUCLEOTIDE COMPLEXES | | Descriptor: | COBALT (II) ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Loll, P.J, Quirk, S, Lattman, E.E. | | Deposit date: | 1994-10-27 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystal structures of staphylococcal nuclease complexed with the competitive inhibitor cobalt(II) and nucleotide.
Biochemistry, 34, 1995
|
|
1TR5
 
 | | Room temperature structure of Staphylococcal nuclease variant truncated Delta+PHS I92E | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Nguyen, D.M, Leila Reynald, R, Gittis, A.G, Lattman, E.E. | | Deposit date: | 2004-06-20 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray and thermodynamic studies of staphylococcal nuclease variants I92E and I92K: insights into polarity of the protein interior
J.Mol.Biol., 341, 2004
|
|
1TT2
 
 | | Cryogenic crystal structure of Staphylococcal nuclease variant truncated Delta+PHS I92K | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Nguyen, D.M, Reynald, R.L, Gittis, A.G, Lattman, E.E. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and thermodynamic studies of staphylococcal nuclease variants I92E and I92K: insights into polarity of the protein interior
J.Mol.Biol., 341, 2004
|
|
1SJY
 
 | | Crystal Structure of NUDIX HYDROLASE DR1025 FROM DEINOCOCCUS RADIODURANS | | Descriptor: | MutT/nudix family protein | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
1SNK
 
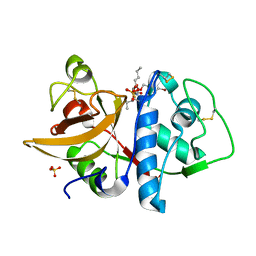 | | Cathepsin K complexed with carbamate derivatized norleucine aldehyde | | Descriptor: | Cathepsin K, N2-({[(4-BROMOPHENYL)METHYL]OXY}CARBONYL)-N1-[(1S)-1-FORMYLPENTYL]-L-LEUCINAMIDE, SULFATE ION | | Authors: | Boros, E.E, Deaton, D.N, Hassell, A.M, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M, Thompson, J.B, Willard Jr, D.H, Wright, L.L. | | Deposit date: | 2004-03-11 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploration of the P(2)-P(3) SAR of aldehyde cathepsin K inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1TQO
 
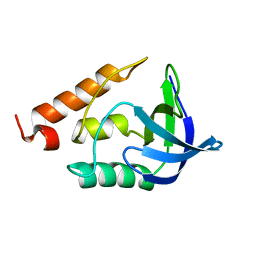 | |
3KDC
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10074 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dichlorophenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chufan, E.E, Kawasaki, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
3KDD
 
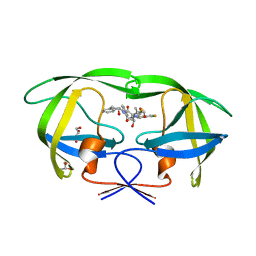 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10265 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-difluorophenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H- inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Chufan, E.E, Kawasaki, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
3LC4
 
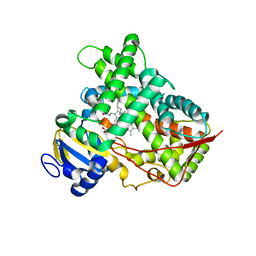 | |
3MID
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide obtained by soaking (100mM NaN3) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3MIH
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide, obtained in the presence of substrate | | Descriptor: | AZIDE ION, COPPER (II) ION, IODIDE ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3JB4
 
 | | Structure of Ljungan virus: insight into picornavirus packaging | | Descriptor: | VP0, VP1, VP3 | | Authors: | Zhu, L, Wang, X.X, Ren, J.S, Porta, C, Wenham, H, Ekstrom, J.-O, Panjwani, A, Knowles, N.J, Kotecha, A, Siebert, A, Lindberg, M, Fry, E.E, Rao, Z.H, Tuthill, T.J, Stuart, D.I. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Ljungan virus provides insight into genome packaging of this picornavirus.
Nat Commun, 6, 2015
|
|
3KDB
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10006 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Chufan, E.E, Lafont, V, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
3MIB
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3MLK
 
 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite | | Descriptor: | COPPER (II) ION, NICKEL (II) ION, NITRITE ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
3MLJ
 
 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound carbon monooxide (CO) | | Descriptor: | ACETATE ION, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Prigge, S.T, Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
3MIG
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite, obtained in the presence of substrate | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
