1U98
 
 | |
6TKX
 
 | | Carbohydrate esterase from gut microbiota | | Descriptor: | Carbohydrate esterase, SULFATE ION | | Authors: | Penttinen, L, Hakulinen, N, Master, R.E. | | Deposit date: | 2019-11-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Polysaccharide utilization loci-driven enzyme discovery reveals BD-FAE: a bifunctional feruloyl and acetyl xylan esterase active on complex natural xylans.
Biotechnol Biofuels, 14, 2021
|
|
1G92
 
 | | SOLUTION STRUCTURE OF PONERATOXIN | | Descriptor: | PONERATOXIN | | Authors: | Szolajska, E, Poznanski, J, Ferber, M.L, Michalik, J, Gout, E, Fender, P, Bailly, I, Dublet, B, Chroboczek, J. | | Deposit date: | 2000-11-22 | | Release date: | 2003-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Poneratoxin, a neurotoxin from ant venom. Structure and expression in insect cells and construction of a bio-insecticide.
Eur.J.Biochem., 271, 2004
|
|
8R2I
 
 | | Cryo-EM Structure of native Photosystem II assembly intermediate from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Fadeeva, M, Klaiman, D, Kandiah, E, Nelson, N. | | Deposit date: | 2023-11-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of native photosystem II assembly intermediate from Chlamydomonas reinhardtii .
Front Plant Sci, 14, 2023
|
|
1GJY
 
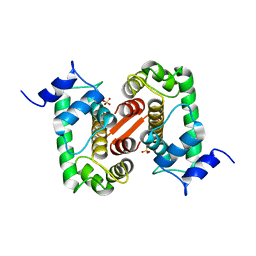 | | The X-ray structure of the Sorcin Calcium Binding Domain (SCBD) provides insight into the phosphorylation and calcium dependent processess | | Descriptor: | SORCIN, SULFATE ION | | Authors: | Ilari, A, Johnson, K.A, Nastopoulos, V, Tsernoglou, D, Chiancone, E. | | Deposit date: | 2001-08-06 | | Release date: | 2002-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Sorcin Calcium Binding Domain Provides a Model of Ca(2+)-Dependent Processes in the Full-Length Protein
J.Mol.Biol., 317, 2002
|
|
1TPL
 
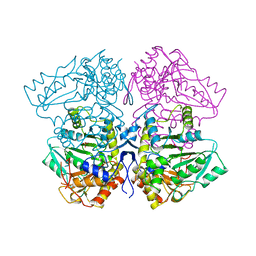 | | THE THREE-DIMENSIONAL STRUCTURE OF TYROSINE PHENOL-LYASE | | Descriptor: | SULFATE ION, TYROSINE PHENOL-LYASE | | Authors: | Antson, A, Demidkina, T, Dauter, Z, Harutyunyan, E, Wilson, K. | | Deposit date: | 1992-11-25 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of tyrosine phenol-lyase.
Biochemistry, 32, 1993
|
|
8QYM
 
 | | Human 20S proteasome assembly intermediate structure 3 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 2024
|
|
8QYN
 
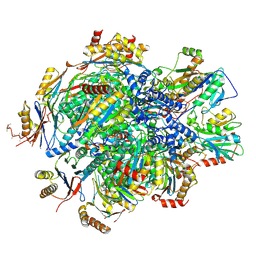 | | Human 20S proteasome assembly intermediate structure 5 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 2024
|
|
8QZ9
 
 | | Human 20S proteasome assembly intermediate structure 4 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 2024
|
|
8QYL
 
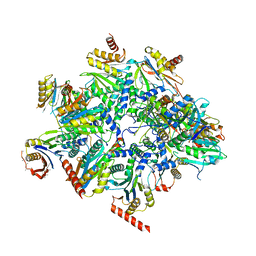 | | Human 20S proteasome assembly intermediate structure 2 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 2024
|
|
8R2C
 
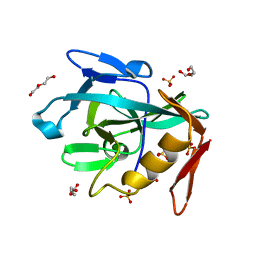 | | Crystal structure of the Vint domain from Tetrahymena thermophila | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, von willebrand factor type A (VWA) domain was originally protein | | Authors: | Iwai, H, Beyer, H.M, Johannson, J.E, Li, M, Wlodawer, A. | | Deposit date: | 2023-11-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of the Vint domain from Tetrahymena thermophila suggests a ligand-regulated cleavage mechanism by the HINT fold.
Febs Lett., 598, 2024
|
|
1FTM
 
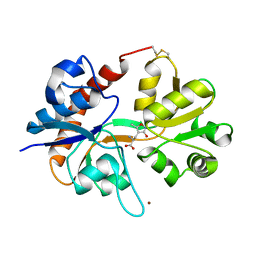 | |
8R10
 
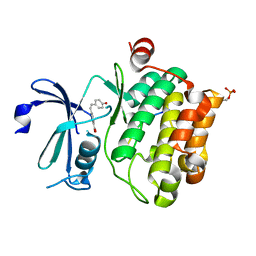 | |
8R1N
 
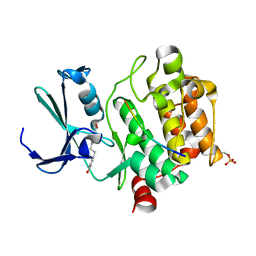 | |
8R1P
 
 | |
6DKB
 
 | | Crystal structure of Trk-A in complex with the Pan-Trk Kinase Inhibitor, compound 10b. | | Descriptor: | 2-{[(3R,4S)-3-fluoro-1-{[4-(trifluoromethoxy)phenyl]acetyl}piperidin-4-yl]oxy}-5-(1-methyl-1H-imidazol-4-yl)pyridine-3-carboxamide, High affinity nerve growth factor receptor | | Authors: | Greasley, S.E, Johnson, E, Kraus, M.L, Cronin, C.N. | | Deposit date: | 2018-05-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of Potent, Selective, and Peripherally Restricted Pan-Trk Kinase Inhibitors for the Treatment of Pain.
J. Med. Chem., 61, 2018
|
|
3FUT
 
 | | Apo-form of T. thermophilus 16S rRNA A1518 and A1519 methyltransferase (KsgA) in space group P21212 | | Descriptor: | Dimethyladenosine transferase | | Authors: | Demirci, H, Belardinelli, R, Seri, E, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5'-methylthioadenosine.
J.Mol.Biol., 388, 2009
|
|
6DKG
 
 | | Crystal structure of Trk-A in complex with the Pan-Trk Kinase Inhibitor, compound 13b. | | Descriptor: | 2-{[(3R,4S)-3-fluoro-1-{[4-(trifluoromethoxy)phenyl]acetyl}piperidin-4-yl]oxy}-4-(2-hydroxy-2-methylpropoxy)benzamide, 5-phenylthieno[2,3-d]pyrimidin-4(3H)-one, High affinity nerve growth factor receptor | | Authors: | Greasley, S.E, Johnson, E, Kraus, M.L, Cronin, C.N. | | Deposit date: | 2018-05-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of Potent, Selective, and Peripherally Restricted Pan-Trk Kinase Inhibitors for the Treatment of Pain.
J. Med. Chem., 61, 2018
|
|
6DKI
 
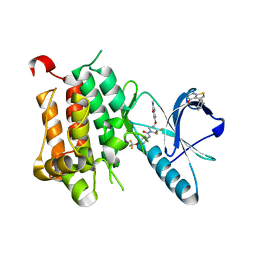 | | Crystal structure of Trk-A in complex with the Pan-Trk Kinase Inhibitor, compound 19. | | Descriptor: | 5-phenylthieno[2,3-d]pyrimidin-4(3H)-one, 6-amino-5-{[(3S)-4,4-difluoro-1-{[4-(trifluoromethoxy)phenyl]acetyl}pyrrolidin-3-yl]oxy}-N-methylpyridine-3-carboxamide, High affinity nerve growth factor receptor | | Authors: | Greasley, S.E, Johnson, E, Kraus, M.L, Cronin, C.N. | | Deposit date: | 2018-05-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of Potent, Selective, and Peripherally Restricted Pan-Trk Kinase Inhibitors for the Treatment of Pain.
J. Med. Chem., 61, 2018
|
|
1G7Y
 
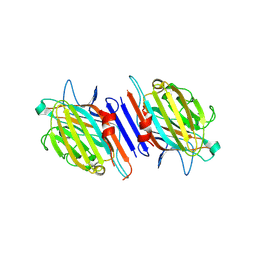 | | THE CRYSTAL STRUCTURE OF THE 58KD VEGETATIVE LECTIN FROM THE TROPICAL LEGUME DOLICHOS BIFLORUS | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, STEM/LEAF LECTIN DB58, ... | | Authors: | Buts, L, Hamelryck, T.W, Loris, R, Dao-Thi, M.-H, Wyns, L, Etzler, M.E. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Weak protein-protein interactions in lectins: the crystal structure of a vegetative lectin from the legume Dolichos biflorus.
J.Mol.Biol., 309, 2001
|
|
3G1H
 
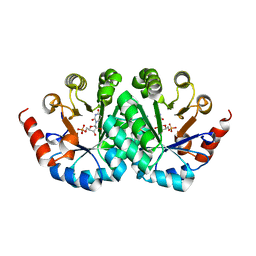 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 5,6-dihydrouridine 5'-monophosphate | | Descriptor: | 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
5VXX
 
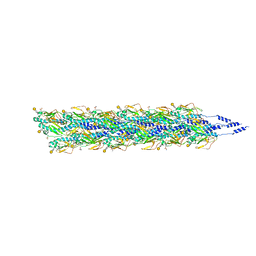 | | Cryo-EM reconstruction of Neisseria gonorrhoeae Type IV pilus | | Descriptor: | Fimbrial protein, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, alpha-D-galactopyranose-(1-3)-2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose | | Authors: | Wang, F, Orlova, A, Altindal, T, Craig, L, Egelman, E.H. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Cryoelectron Microscopy Reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae Type IV Pili at Sub-nanometer Resolution.
Structure, 25, 2017
|
|
8R1T
 
 | | Pim1 in complex with 4-(4-aminophenethyl)benzoic acid and Pimtide | | Descriptor: | 4-(4-aminophenethyl)benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2023-11-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | What doesn't fit is made to fit: Pim-1 kinase adapts to the configuration of stilbene-based inhibitors.
Arch Pharm, 357, 2024
|
|
1UIJ
 
 | | Crystal Structure Of Soybean beta-Conglycinin Beta Homotrimer (I122M/K124W) | | Descriptor: | beta subunit of beta conglycinin | | Authors: | Maruyama, N, Maruyama, Y, Tsuruki, T, Okuda, E, Yoshikawa, M, Utsumi, S. | | Deposit date: | 2003-07-16 | | Release date: | 2004-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Creation of soybean beta-conglycinin beta with strong phagocytosis-stimulating activity
BIOCHIM.BIOPHYS.ACTA, 1648, 2003
|
|
6TQJ
 
 | | Crystal structure of the c14 ring of the F1FO ATP synthase from spinach chloroplast | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ATP synthase subunit c, chloroplastic, ... | | Authors: | Kovalev, K, Gushchin, I, Vlasov, A, Round, E, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2019-12-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unusual features of the c-ring of F1FOATP synthases.
Sci Rep, 9, 2019
|
|
