7AV8
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212 | | Descriptor: | PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
To Be Published
|
|
1UXN
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-02-27 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
6SPR
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase variant VI298 complexed with beta-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
5J01
 
 | | Structure of the lariat form of a chimeric derivative of the Oceanobacillus iheyensis group II intron in the presence of NH4+ and MG2+. | | Descriptor: | AMMONIUM ION, MAGNESIUM ION, group II intron lariat | | Authors: | Costa, M, Walbott, H, Monachello, D, Westhof, E, Michel, F. | | Deposit date: | 2016-03-26 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Crystal structures of a group II intron lariat primed for reverse splicing.
Science, 354, 2016
|
|
1UZU
 
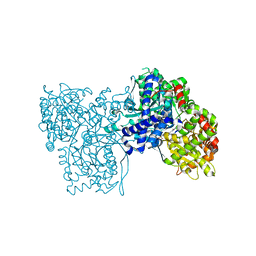 | | Glycogen Phosphorylase b in complex with indirubin-5'-sulphonate | | Descriptor: | 2',3-DIOXO-1,1',2',3-TETRAHYDRO-2,3'-BIINDOLE-5'-SULFONIC ACID, GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, ... | | Authors: | Kosmopoulou, M.N, Leonidas, D.D, Chrysina, E.D, Bischler, N, Eisenbrand, G, Sakarellos, C.E, Pauptit, R, Oikonomakos, N.G. | | Deposit date: | 2004-03-16 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of the potential antitumour agent indirubin-5-sulphonate at the inhibitor site of rabbit muscle glycogen phosphorylase b. Comparison with ligand binding to pCDK2-cyclin A complex.
Eur. J. Biochem., 271, 2004
|
|
6F5D
 
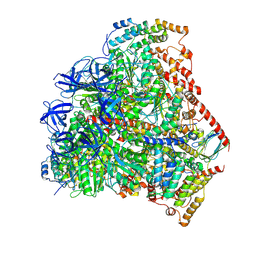 | | Trypanosoma brucei F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase gamma subunit, ATP synthase subunit alpha, ... | | Authors: | Montgomery, M.G, Gahura, O, Leslie, A.G.W, Zikova, A, Walker, J.E. | | Deposit date: | 2017-12-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | ATP synthase fromTrypanosoma bruceihas an elaborated canonical F1-domain and conventional catalytic sites.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6SES
 
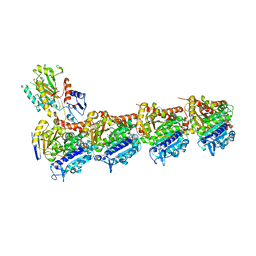 | | Tubulin-B2 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Guo, B, Rodriguez-Gabin, A, Prota, A.E, Muehlethaler, T, Zhang, N, Ye, K, Steinmetz, M.O, Band Horwitz, S, Smith III, A.B, McDaid, H.M. | | Deposit date: | 2019-07-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Refinement of the Tubulin Ligand (+)-Discodermolide to Attenuate Chemotherapy-Mediated Senescence.
Mol.Pharmacol., 98, 2020
|
|
6B04
 
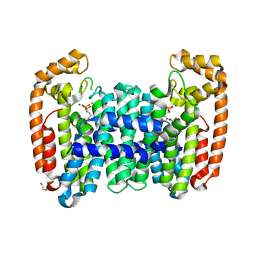 | | Crystal structure of CfFPPS2, a lepidopteran type-II farnesyl diphosphate synthase, complexed with [2-(1-methylpyridin-2-yl)-1-phosphono-ethyl]phosphonic acid (inhibitor 1b) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,2-diphosphonoethyl)-1-methylpyridin-1-ium, Farnesyl diphosphate synthase, ... | | Authors: | Picard, M.-E, Cusson, M, Shi, R. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural characterization of a lepidopteran type-II farnesyl diphosphate synthase from the spruce budworm, Choristoneura fumiferana: Implications for inhibitor design.
Insect Biochem. Mol. Biol., 92, 2017
|
|
7NCT
 
 | | Glutathione-S-transferase GliG mutant K127G | | Descriptor: | Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7WTL
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Dis-D | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, La Venuta, G, Lau, B, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In vitro structural maturation of an early stage pre-40S particle coupled with U3 snoRNA release and central pseudoknot formation.
Nucleic Acids Res., 50, 2022
|
|
7NCE
 
 | | Glutathione-S-transferase GliG mutant N27A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6YO1
 
 | | Crystal structure of ribonuclease A solved by vanadium SAD phasing | | Descriptor: | Ribonuclease pancreatic, URIDINE-2',3'-VANADATE | | Authors: | El Omari, K, Mohamad, N, Bountra, K, Duman, R, Romano, M, Schlegel, K, Kwong, H, Mykhaylyk, V, Olesen, C.E, Moller, J.V, Bublitz, M, Beis, K, Wagner, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental phasing with vanadium and application to nucleotide-binding membrane proteins.
Iucrj, 7, 2020
|
|
7NCL
 
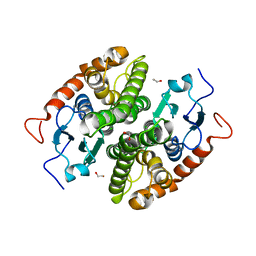 | | Glutathione-S-transferase GliG mutant E82Q | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6F86
 
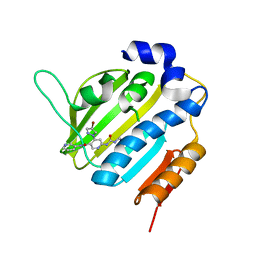 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 4-(4-bromo-1H-pyrazol-1-yl)-6-[(ethylcarbamoyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 4-(4-bromanylpyrazol-1-yl)-6-(ethylcarbamoylamino)-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
2IF7
 
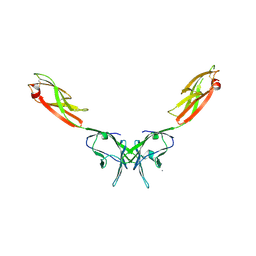 | | Crystal Structure of NTB-A | | Descriptor: | CALCIUM ION, CHLORIDE ION, SLAM family member 6 | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2006-09-20 | | Release date: | 2006-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | NTB-A Receptor Crystal Structure: Insights into Homophilic Interactions in the Signaling Lymphocytic Activation Molecule Receptor Family.
Immunity, 25, 2006
|
|
7NCD
 
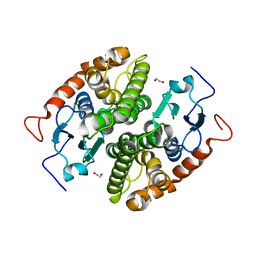 | | Glutathione-S-transferase GliG mutant N27D | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glutathione S-transferase GliG, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC8
 
 | | Glutathione-S-transferase GliG mutant S24A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4P6X
 
 | | Crystal Structure of cortisol-bound glucocorticoid receptor ligand binding domain | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
6IA7
 
 | | T. brucei IFT22 GTP-bound crystal structure | | Descriptor: | CALCIUM ION, GUANOSINE-5'-TRIPHOSPHATE, Intraflagellar transport protein 22, ... | | Authors: | Wachter, S, Basquin, J, Lorentzen, E. | | Deposit date: | 2018-11-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of IFT22 to the intraflagellar transport complex is essential for flagellum assembly.
Embo J., 38, 2019
|
|
7NCN
 
 | | Glutathione-S-transferase GliG mutant S83A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6IAE
 
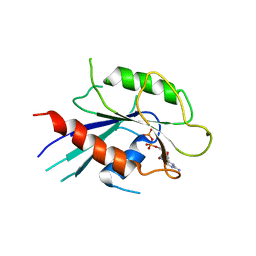 | | T. brucei IFT22 GDP-bound crystal structure | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Intraflagellar transport protein 22, MAGNESIUM ION | | Authors: | Wachter, S, Basquin, J, Lorentzen, E. | | Deposit date: | 2018-11-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Binding of IFT22 to the intraflagellar transport complex is essential for flagellum assembly.
Embo J., 38, 2019
|
|
5FYK
 
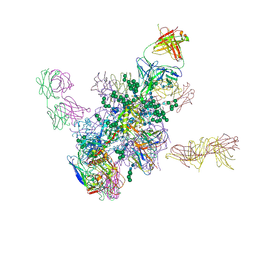 | | Crystal Structure at 3.7 A Resolution of Fully Glycosylated HIV-1 Clade B JR-FL SOSIP.664 Prefusion Env Trimer in Complex with Broadly Neutralizing Antibodies PGT122, 35O22 and VRC01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Thomas, P.V, Kwong, P.D. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.107 Å) | | Cite: | Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G
Cell(Cambridge,Mass.), 165, 2016
|
|
7NC3
 
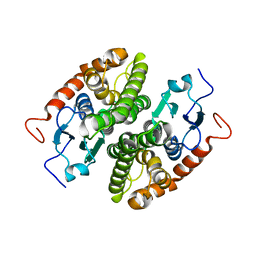 | |
2IHY
 
 | | Structure of the Staphylococcus aureus putative ATPase subunit of an ATP-binding cassette (ABC) transporter | | Descriptor: | ABC transporter, ATP-binding protein, SULFATE ION | | Authors: | McGrath, T.E, Yu, C.S, Romanov, V, Lam, R, Dharamsi, A, Virag, C, Mansoury, K, Thambipillai, D, Richards, D, Guthrie, J, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2006-09-27 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Staphylococcus aureus putative ATPase subunit of an ATP-binding cassette (ABC) transporter
To be Published
|
|
6SAV
 
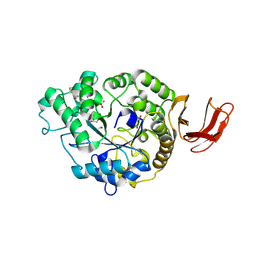 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, CALCIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
