5VOV
 
 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Zhao, Y, Chen, S, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
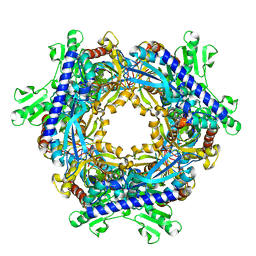
5HKK
 
 | | Caldalaklibacillus thermarum F1-ATPase (wild type) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Ferguson, S.A, Cook, G.M, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1T5G
 
 | | Arginase-F2-L-Arginine complex | | Descriptor: | ARGININE, Arginase 1, FLUORIDE ION, ... | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Han, S, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-05-04 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
3KLH
 
 | | Crystal structure of AZT-Resistant HIV-1 Reverse Transcriptase crosslinked to post-translocation AZTMP-Terminated DNA (COMPLEX P) | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*T*GP*CP*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), MAGNESIUM ION, ... | | Authors: | Tu, X, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6GQ3
 
 | | Human asparagine synthetase (ASNS) in complex with 6-diazo-5-oxo-L-norleucine (DON) at 1.85 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-OXO-L-NORLEUCINE, ... | | Authors: | Zhu, W, Radadiya, A, Bisson, C, Jin, Y, Nordin, B.E, Imasaki, T, Wenzel, S, Sedelnikova, S.E, Berry, A.H, Nomanbhoy, T.K, Kozarich, J.W, Takagi, Y, Rice, D.W, Richards, N.G.J. | | Deposit date: | 2018-06-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-resolution crystal structure of human asparagine synthetase enables analysis of inhibitor binding and selectivity.
Commun Biol, 2, 2019
|
|
1M3V
 
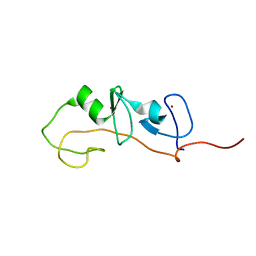 | | FLIN4: Fusion of the LIM binding domain of Ldb1 and the N-terminal LIM domain of LMO4 | | Descriptor: | ZINC ION, fusion of the LIM interacting domain of ldb1 and the N-terminal LIM domain of LMO4 | | Authors: | Deane, J.E, Mackay, J.P, Kwan, A.H.Y, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2002-06-30 | | Release date: | 2003-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
1T8S
 
 | |
1T9O
 
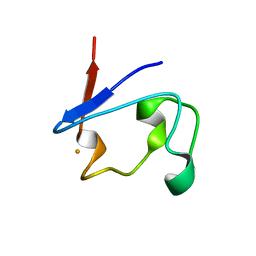 | | Crystal Structure of V44G Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
5HNO
 
 | | The structure of the kdo-capped saccharide binding subunit of the O-12 specific ABC transporter, Wzt | | Descriptor: | ABC type transport system putative ATP binding protein, CHLORIDE ION | | Authors: | Mallette, E, Mann, E, Whitfield, C, Kimber, M.S. | | Deposit date: | 2016-01-18 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Klebsiella pneumoniae O12 ATP-binding Cassette (ABC) Transporter Recognizes the Terminal Residue of Its O-antigen Polysaccharide Substrate.
J.Biol.Chem., 291, 2016
|
|
1TA1
 
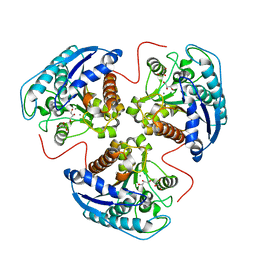 | | H141C mutant of rat liver arginase I | | Descriptor: | Arginase 1, GLYCEROL, MANGANESE (II) ION | | Authors: | Cama, E, Cox, J.D, Ash, D.E, Christianson, D.W. | | Deposit date: | 2004-05-19 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
5H6Z
 
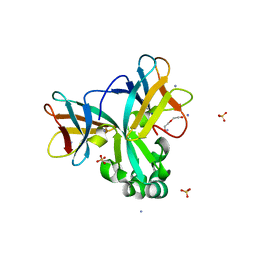 | | Crystal structure of Set7, a novel histone methyltransferase in Schizossacharomyces pombe | | Descriptor: | AMMONIUM ION, DI(HYDROXYETHYL)ETHER, SET domain-containing protein 7, ... | | Authors: | Mevius, D.E.H.F, Shen, Y, Morishita, M, Carrozzini, B, Caliandro, R, di Luccio, E. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Set7 Is a H3K37 Methyltransferase in Schizosaccharomyces pombe and Is Required for Proper Gametogenesis.
Structure, 27, 2019
|
|
1BDV
 
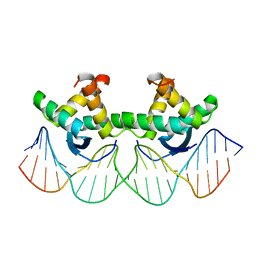 | | ARC FV10 COCRYSTAL | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*AP*TP*AP*GP*AP*AP*GP*CP*AP*CP*TP*CP*TP*AP*CP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*GP*TP*AP*GP*AP*GP*TP*GP*CP*TP*TP*CP*TP*AP*TP*CP*AP*T)-3'), PROTEIN (ARC FV10 REPRESSOR) | | Authors: | Schildbach, J.F, Karzai, A.W, Raumann, B.E, Sauer, R.T. | | Deposit date: | 1998-05-11 | | Release date: | 1999-01-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Origins of DNA-binding specificity: role of protein contacts with the DNA backbone.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7KXS
 
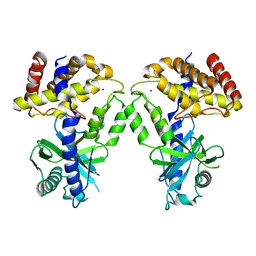 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2020-12-04 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5VWW
 
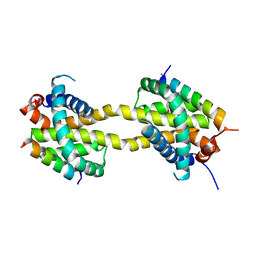 | | Bak core latch dimer in complex with Bim-RT - Tetragonal | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BROMIDE ION, Bcl-2 homologous antagonist/killer, ... | | Authors: | Brouwer, J.M, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Conversion of Bim-BH3 from Activator to Inhibitor of Bak through Structure-Based Design.
Mol. Cell, 68, 2017
|
|
5W3F
 
 | | Yeast tubulin polymerized with GTP in vitro | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Howes, S.C, Geyer, E.A, LaFrance, B, Zhang, R, Kellogg, E.H, Westermann, S, Rice, L.M, Nogales, E. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural differences between yeast and mammalian microtubules revealed by cryo-EM.
J. Cell Biol., 216, 2017
|
|
1TGV
 
 | | Structure of E. coli Uridine Phosphorylase complexed with 5-Fluorouridine and sulfate | | Descriptor: | 5-FLUOROURIDINE, POTASSIUM ION, SULFATE ION, ... | | Authors: | Bu, W, Settembre, E.C, Sanders, J.M, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-05-31 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of E. coli Uridine Phosphorylase
To be Published, 2004
|
|
6CUT
 
 | | Engineered Holo TrpB from Pyrococcus furiosus, PfTrpB7E6 with (2S,3S)-isopropylserine bound as the external aldimine | | Descriptor: | (2S,3S)-3-hydroxy-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-methylpentanoic acid (non-preferred name), SODIUM ION, Tryptophan synthase beta chain 1 | | Authors: | Boville, C.E, Scheele, R.A, Buller, A.R, Arnold, F.H. | | Deposit date: | 2018-03-26 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Engineered Biosynthesis of beta-Alkyl Tryptophan Analogues.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6CVJ
 
 | |
4QR9
 
 | | Crystal structure of two HMGB1 Box A domains cooperating to underwind and kink a DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*GP*AP*TP*AP*T)-3'), High mobility group protein B1, MAGNESIUM ION | | Authors: | Sanchez-Giraldo, R, Acosta-Reyes, F.J, Malarkey, C.S, Saperas, N, Churchill, M.E.A, Campos, J.L. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two high-mobility group box domains act together to underwind and kink DNA.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6CY6
 
 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Escherichia coli in complex with tris(hydroxymethyl)aminomethane. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-04 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1T5Z
 
 | | Crystal Structure of the Androgen Receptor Ligand Binding Domain (LBD) with DHT and a peptide derived from its physiological coactivator ARA70 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 4 | | Authors: | Estebanez-Perpina, E, Moore, J.M.R, Mar, E, Nguyen, P, Delgado-Rodrigues, E, Baxter, J.D, Webb, P, Fletterick, R.J, Guy, R.K. | | Deposit date: | 2004-05-05 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Molecular Mechanisms of Coactivator Utilization in Ligand-dependent Transactivation by the Androgen Receptor.
J.Biol.Chem., 280, 2005
|
|
1T63
 
 | | Crystal Structure of the Androgen Receptor Ligand Binding Domain with DHT and a peptide derived from its physiological coactivator GRIP1 NR box3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 2 | | Authors: | Estebanez-Perpina, E, Moore, J.M.R, Mar, E, Nguyen, P, Delgado-Rodrigues, E, Baxter, J.D, Webb, P, Fletterick, R.J, Guy, R.K. | | Deposit date: | 2004-05-05 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Molecular Mechanisms of Coactivator Utilization in Ligand-dependent Transactivation by the Androgen Receptor.
J.Biol.Chem., 280, 2005
|
|
7ZPP
 
 | | Cryo-EM structure of the MVV CSC intasome at 4.5A resolution | | Descriptor: | Integrase, vDNA, non-transferred strand, ... | | Authors: | Ballandras-Colas, A, Maskell, D, Pye, V.E, Locke, J, Swuec, S, Kotecha, A, Costa, A, Cherepanov, P. | | Deposit date: | 2022-04-28 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A supramolecular assembly mediates lentiviral DNA integration
Science, 355, 2017
|
|
7ZBP
 
 | | Unspecific peroxygenase from Marasmius rotula | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2022-03-24 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Characterization of Two Short Unspecific Peroxygenases: Two Different Dimeric Arrangements.
Antioxidants, 11, 2022
|
|
7ZCL
 
 | | Unspecific peroxygenase from Collariella virescens | | Descriptor: | Collariella virescens UPO, HEME C, MAGNESIUM ION | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2022-03-28 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization of Two Short Unspecific Peroxygenases: Two Different Dimeric Arrangements.
Antioxidants, 11, 2022
|
|
