8AXE
 
 | | Small molecule stabilizer for ERalpha and 14-3-3 (1074210) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[2-[1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]piperidin-4-yl]ethyl]ethanamide, Estrogen receptor, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-08-31 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AXU
 
 | | Small molecule stabilizer for ERalpha and 14-3-3 (1075297) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[1-(4-chloranylphenoxy)cyclopentyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8ATP
 
 | | Small molecule stabilizer (1075481) for ERalpha and 14-3-3 | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[4-[(4-chlorophenyl)amino]piperidin-4-yl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Vandenboorn, E.M.F, Visser, E.J, Ottmann, C. | | Deposit date: | 2022-08-23 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AUS
 
 | | Small molecular stabilizer for ERalpha and 14-3-3 (1080297) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[7-[4-[(4-chlorophenyl)amino]oxan-4-yl]carbonyl-7-azaspiro[3.5]nonan-2-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-25 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AV4
 
 | | Small molecular stabilizer for ERalpha and 14-3-3 (1075305) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[4-(4-chloranylphenoxy)oxan-4-yl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-26 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
7PSX
 
 | | Structure of HOXB13 bound to hydroxymethylated DNA | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*TP*5HCP*AP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*CP*GP*AP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13, ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2021-09-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of HOXB13 bound to hydroxymethylated DNA
To Be Published
|
|
8V6G
 
 | | DNA initiation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.16 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6H
 
 | | DNA initiation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.11 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7NWT
 
 | | Initiated 70S ribosome in complex with 2A protein from encephalomyocarditis virus (EMCV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Hill, C.H, Napthine, S, Pekarek, L, Kibe, A, Firth, A.E, Graham, S.C, Caliskan, N, Brierley, I. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-15 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural and molecular basis for Cardiovirus 2A protein as a viral gene expression switch
Nat Commun, 12, 2021
|
|
6X80
 
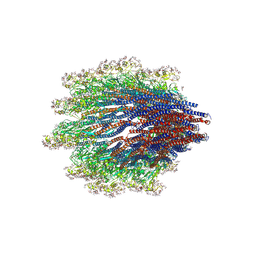 | | Structure of the Campylobacter jejuni G508A Flagellar Filament | | Descriptor: | 5,7-diamino-3,5,7,9-tetradeoxy-L-glycero-alpha-L-manno-non-2-ulopyranosonic acid, Flagellin A | | Authors: | Kreutzberger, M.A.B, Wang, F, Egelman, E.H. | | Deposit date: | 2020-06-01 | | Release date: | 2020-07-08 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structure of the Campylobacter jejuni flagellar filament reveals how epsilon Proteobacteria escaped Toll-like receptor 5 surveillance.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4F
 
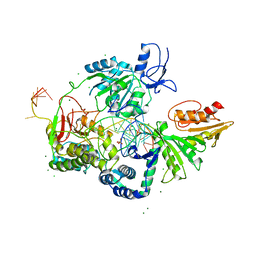 | | Crystal structure of the XPB-Bax1-forked DNA ternary complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*AP*AP*CP*AP*TP*CP*CP*TP*TP*TP*GP*CP*TP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*GP*TP*AP*GP*GP*TP*TP*TP*CP*CP*AP*TP*GP*TP*TP*GP*AP*GP*TP*CP*A)-3'), ... | | Authors: | He, F, Hilario, E, Fan, L. | | Deposit date: | 2019-05-27 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural basis of the XPB helicase-Bax1 nuclease complex interacting with the repair bubble DNA.
Nucleic Acids Res., 48, 2020
|
|
4XDS
 
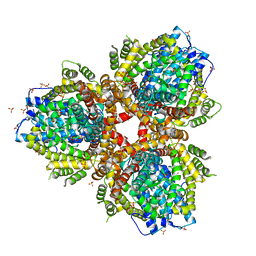 | | Deoxyguanosinetriphosphate Triphosphohydrolase from Escherichia coli with Nickel | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, NICKEL (II) ION, SULFATE ION | | Authors: | Singh, D, Gawel, D, Itsko, M, Krahn, J.M, London, R.E, Schaaper, R.M. | | Deposit date: | 2014-12-19 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | Structure of Escherichia coli dGTP Triphosphohydrolase: A HEXAMERIC ENZYME WITH DNA EFFECTOR MOLECULES.
J.Biol.Chem., 290, 2015
|
|
6X9F
 
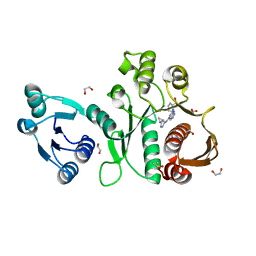 | | Pseudomonas aeruginosa MurC with AZ8074 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Horanyi, P.S, Mayclin, S.J, Durand-Reville, T.F, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Pseudomonas aeruginosa MurC with AZ8074
To Be Published
|
|
7NGJ
 
 | |
7NGD
 
 | |
7NGS
 
 | |
7NGX
 
 | |
7NGO
 
 | |
7NGK
 
 | |
7NGT
 
 | |
7NGN
 
 | |
7NGR
 
 | |
7NGG
 
 | |
7NGM
 
 | |
7NGI
 
 | |
