7NKA
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8206 | | Descriptor: | NB8206, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIL
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8190 core | | Descriptor: | Nanobody8190 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.01 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIR
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8191 core | | Descriptor: | Nanobody8191 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-13 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NK2
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8202 core | | Descriptor: | Nanobody8202, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.84 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NJ4
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8198 core | | Descriptor: | Nb8198 Core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.84 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NK4
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8203 core | | Descriptor: | Nanobody 8203, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.32 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NJ3
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8196 core | | Descriptor: | Nanobody8196 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.48 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NK8
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8205 core | | Descriptor: | Nb8205, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.34 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7O3U
 
 | | The crystal structure of obelin from Obelia longissima bound with v-coelenterazine | | Descriptor: | CALCIUM ION, Obelin, v-coelenterazine | | Authors: | Larionova, M.D, Wu, L.J, Vysotski, E.S, Liu, Z.-J. | | Deposit date: | 2021-04-03 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of semisynthetic obelin-v.
Protein Sci., 31, 2022
|
|
7NGU
 
 | |
7NGY
 
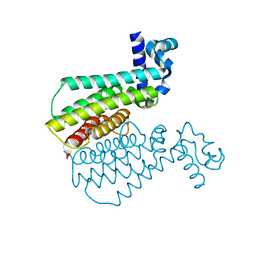 | |
7NGW
 
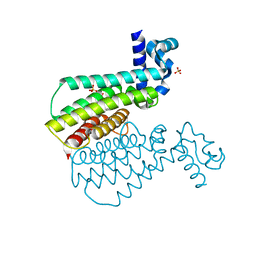 | |
1UGO
 
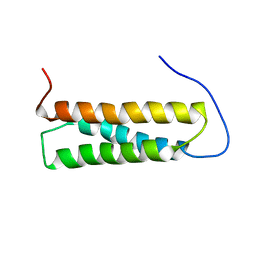 | | Solution structure of the first Murine BAG domain of Bcl2-associated athanogene 5 | | Descriptor: | Bcl2-associated athanogene 5 | | Authors: | Endoh, H, Hayashi, F, Seimiya, K, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-17 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
6PE2
 
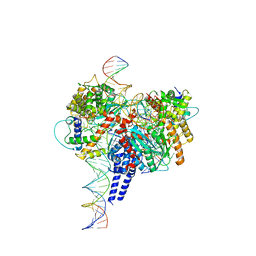 | | Drosophila P element transposase strand transfer complex | | Descriptor: | DNA (27-MER), DNA (5'-D(P*CP*GP*AP*AP*CP*TP*AP*TP*A)-3'), DNA (56-MER), ... | | Authors: | Kellogg, E.H, Nogales, E, Ghanim, G, Rio, D.C. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of a P element transposase-DNA complex reveals unusual DNA structures and GTP-DNA contacts.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6FQV
 
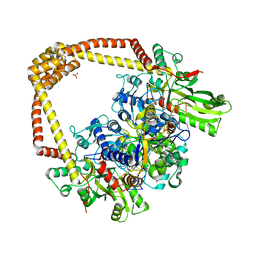 | | 2.60A BINARY COMPLEX OF S.AUREUS GYRASE with UNCLEAVED DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*GP*TP*AP*CP*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, DNA gyrase subunit B,DNA gyrase subunit B, ... | | Authors: | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
8BXU
 
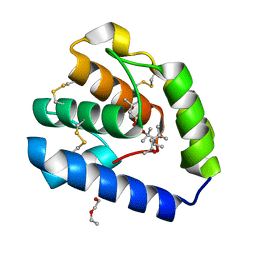 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with MPD (2-Methyl-2,4-pentanediol) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-ETHOXYETHANOL, Odorant binding protein, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-09 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
8BXV
 
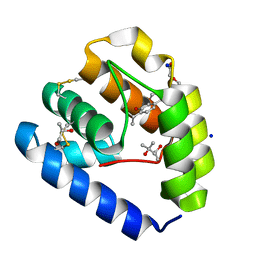 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with Thymol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-METHYL-2-(1-METHYLETHYL)PHENOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
8BN6
 
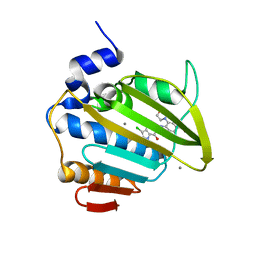 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with EBL3021 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-morpholin-4-yl-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, DNA gyrase subunit B | | Authors: | Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2022-11-12 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Dual Inhibitors of Bacterial Topoisomerases with Broad-Spectrum Antibacterial Activity and In Vivo Efficacy against Vancomycin-Intermediate Staphylococcus aureus .
J.Med.Chem., 66, 2023
|
|
6ZZY
 
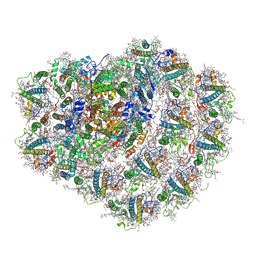 | | Structure of high-light grown Chlorella ohadii photosystem I | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ... | | Authors: | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | Deposit date: | 2020-08-05 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
7A4P
 
 | | Structure of small high-light grown Chlorella ohadii photosystem I | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | Authors: | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-07-28 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
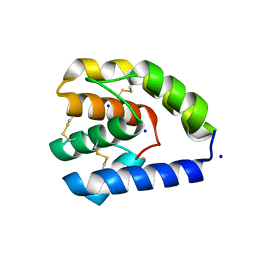
8C6G
 
 | |
8C68
 
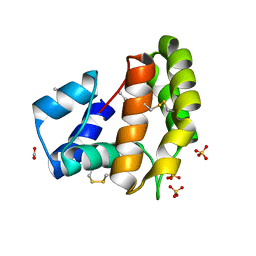 | |
7OXA
 
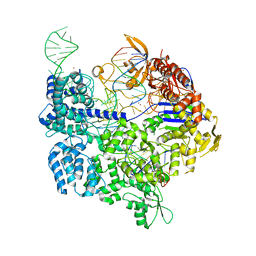 | | Target-bound SpCas9 complex with AAVS1 chimeric RNA-DNA guide | | Descriptor: | AAVS1 non-target DNA strand, AAVS1 target DNA strand, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OX7
 
 | | Target-bound SpCas9 complex with TRAC chimeric RNA-DNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OX8
 
 | | Target-bound SpCas9 complex with TRAC full RNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
