7OTN
 
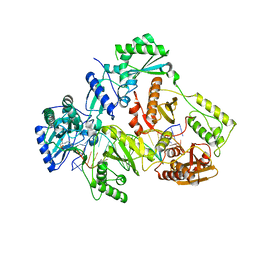 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-247 | | Descriptor: | (S)-2-((2-(6-amino-9H-purin-9-yl)ethyl)amino)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OZ2
 
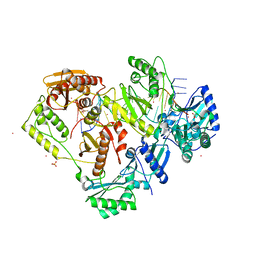 | | Crystal structure of HIV-1 reverse transcriptase with a double stranded DNA showing a transient P-pocket | | Descriptor: | CADMIUM ION, DNA (28-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Das, K. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Sliding of HIV-1 reverse transcriptase over DNA creates a transient P pocket - targeting P-pocket by fragment screening.
Nat Commun, 12, 2021
|
|
7OTZ
 
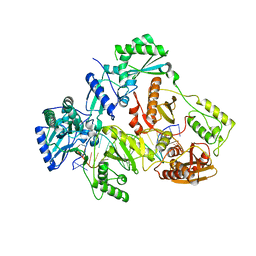 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-259 | | Descriptor: | (S)-2-(2-(6-amino-9H-purin-9-yl)ethoxy)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OOT
 
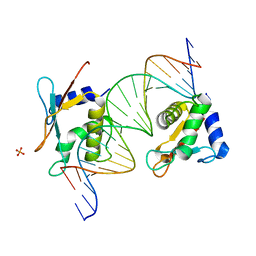 | | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to an interferon-stimulated response element | | Descriptor: | DNA (5'-D(P*AP*GP*CP*TP*TP*TP*CP*TP*CP*GP*GP*TP*TP*TP*CP*AP*GP*TP*TP*G)-3'), DNA (5'-D(P*TP*CP*AP*AP*CP*TP*GP*AP*AP*AP*CP*CP*GP*AP*GP*AP*AP*AP*GP*C)-3'), Interferon regulatory factor 4, ... | | Authors: | Agnarelli, A, El Omari, K, Alt, A.O, Mancini, E.J. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to interferon-stimulated response element
To Be Published
|
|
9NX6
 
 | | Human GAR transformylase in complex with GAR substrate and AGF302 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[4-(2-amino-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]-3-fluorothiophene-2-carbonyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2025-03-25 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Multitargeted 6-Substituted Thieno[2,3- d ]pyrimidines as Folate Receptor-Selective Anticancer Agents that Inhibit Cytosolic and Mitochondrial One-Carbon Metabolism.
Acs Pharmacol Transl Sci, 6, 2023
|
|
9O58
 
 | | Zymogen ADAM17-iRhom2 complex bound by the MEDI3622 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Slone, C.E, Maciag, J.J. | | Deposit date: | 2025-04-09 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural insights into the activation and inhibition of the ADAM17-iRhom2 complex.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
7NOS
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with 4-bromo-6-(trifluoromethyl)-1H-benzo[d]imidazole. | | Descriptor: | 4-bromanyl-6-(trifluoromethyl)-1~{H}-benzimidazole, Ornithine carbamoyltransferase, PHOSPHATE ION | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
9QEK
 
 | | Structure of Human ROS1 Kinase Domain Harboring the G2032R Solvent-front Mutation in Complex with Zidesamtinib (NVL-520) | | Descriptor: | 1,2-ETHANEDIOL, Proto-oncogene tyrosine-protein kinase ROS, Zidesamtinib | | Authors: | Tangpeerachaikul, A, Mente, S, Magrino, J, Gu, F, Horan, J.C, Pelish, H.E. | | Deposit date: | 2025-03-10 | | Release date: | 2025-04-30 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Zidesamtinib Selective Targeting of Diverse ROS1 Drug-Resistant Mutations.
Mol.Cancer Ther., 24, 2025
|
|
7NNZ
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with 5-methyl-4-phenylthiazol-2-amine. | | Descriptor: | 5-methyl-4-phenyl-1,3-thiazol-2-amine, Ornithine carbamoyltransferase, PHOSPHATE ION | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNV
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with carbamoyl phosphate. | | Descriptor: | 1,2-ETHANEDIOL, Ornithine carbamoyltransferase, PHOSPHATE ION, ... | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NP0
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with (4-nitrophenyl)boronic acid. | | Descriptor: | Ornithine carbamoyltransferase, PHOSPHATE ION, p-nitrophenylboronic acid | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NOV
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with (4-methyl-3-nitrophenyl)boronic acid. | | Descriptor: | (4-methyl-3-nitro-phenyl)-oxidanyl-oxidanylidene-boron, Ornithine carbamoyltransferase, PHOSPHATE ION | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7O0H
 
 | |
9JUR
 
 | |
9R2K
 
 | |
7OF2
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with GTPBP6. | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
7OF4
 
 | | Structure of mature human mitochondrial ribosome large subunit in complex with GTPBP6 (PTC conformation 1). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
7OF6
 
 | | Structure of mature human mitochondrial ribosome large subunit in complex with GTPBP6 (PTC conformation 2). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
7OF5
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 and GTPBP5 (dataset2). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
7OF0
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 (dataset1). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
7OG3
 
 | | IRED-88 | | Descriptor: | NAD(P)-dependent oxidoreductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION | | Authors: | Faller, M, Koch, E. | | Deposit date: | 2021-05-06 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Machine-Directed Evolution of an Imine Reductase for Activity and Stereoselectivity
Acs Catalysis, 2021
|
|
7NOU
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with (3,5-dichlorophenyl)boronic acid. | | Descriptor: | Ornithine carbamoyltransferase, PHOSPHATE ION, [3,5-bis(chloranyl)phenyl]-oxidanyl-oxidanylidene-boron | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNF
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in apo form. | | Descriptor: | Ornithine carbamoyltransferase, PHOSPHATE ION | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NIY
 
 | | E. coli NfsA with FMN | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Day, M.D, Jarrom, D, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-14 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
7NNX
 
 | | E. coli NfsA with 1,4-benzoquinone | | Descriptor: | 1,2-ETHANEDIOL, 1,4-benzoquinone, CHLORIDE ION, ... | | Authors: | Day, M.D, Jarrom, D, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
