1MN6
 
 | | Thioesterase Domain from Picromycin Polyketide Synthase, pH 7.6 | | Descriptor: | polyketide synthase IV | | Authors: | Tsai, S.-C, Lu, H, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-09-05 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases
Biochemistry, 41, 2002
|
|
9CC8
 
 | | Hexameric state of the NRC4 resistosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, NLR-required for cell death 4 | | Authors: | Liu, F, Yang, Z, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2024-06-21 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Activation of the helper NRC4 immune receptor forms a hexameric resistosome.
Cell, 187, 2024
|
|
9CC9
 
 | | Dodecameric state of the NRC4 resistosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, NLR-required for cell death 4 | | Authors: | Liu, F, Yang, Z, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2024-06-21 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Activation of the helper NRC4 immune receptor forms a hexameric resistosome.
Cell, 187, 2024
|
|
9BXI
 
 | |
9CJ0
 
 | |
8TF3
 
 | | Wildtype rabbit TRPV5 into nanodiscs in complex with econazole | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[(2S)-2-[(4-CHLOROBENZYL)OXY]-2-(2,4-DICHLOROPHENYL)ETHYL]-1H-IMIDAZOLE, ERGOSTEROL, ... | | Authors: | De Jesus-Perez, J.J, Fluck, E.C, Moiseenkova-Bell, V.Y. | | Deposit date: | 2023-07-07 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural mechanism of TRPV5 inhibition by econazole.
Structure, 32, 2024
|
|
8TF2
 
 | |
8TF4
 
 | | Wildtype rabbit TRPV5 into nanodiscs in the presence of PI(4,5)P2 and econazole | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[(2S)-2-[(4-CHLOROBENZYL)OXY]-2-(2,4-DICHLOROPHENYL)ETHYL]-1H-IMIDAZOLE, ERGOSTEROL, ... | | Authors: | De Jesus-Perez, J.J, Fluck, E.C, Moiseenkova-Bell, V.Y. | | Deposit date: | 2023-07-07 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural mechanism of TRPV5 inhibition by econazole.
Structure, 32, 2024
|
|
9BXO
 
 | |
8TVP
 
 | | Cryo-EM structure of CPD-stalled Pol II in complex with Rad26 (open state) | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVV
 
 | | Cryo-EM structure of backtracked Pol II | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVQ
 
 | | Cryo-EM structure of CPD stalled 10-subunit Pol II in complex with Rad26 | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVX
 
 | | Cryo-EM structure of CPD-stalled Pol II (Conformation 2) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVY
 
 | | Cryo-EM structure of CPD lesion containing RNA Polymerase II elongation complex with Rad26 and Elf1 (closed state) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVW
 
 | | Cryo-EM structure of CPD-stalled Pol II (conformation 1) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVS
 
 | | Cryo-EM structure of backtracked Pol II in complex with Rad26 | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TUG
 
 | | Cryo-EM structure of CPD-stalled Pol II in complex with Rad26 (engaged state) | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-16 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9G9S
 
 | | Crystal structure of PbdA bound to veratrate | | Descriptor: | 3,4-dimethoxybenzoic acid, Cytochrome P450 CYP199, GLYCEROL, ... | | Authors: | Hinchen, D.J, Wolf, M.E, Eltis, L.D, McGeehan, J.E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of a cytochrome P450 that catalyzes the O-demethylation of lignin-derived benzoates.
J.Biol.Chem., 2024
|
|
9G9Q
 
 | | Crystal structure of PbdA bound to p-methoxybenzoate. | | Descriptor: | 4-METHOXYBENZOIC ACID, Cytochrome P450 CYP199, GLYCEROL, ... | | Authors: | Hinchen, D.J, Wolf, M.E, Eltis, L.D, McGeehan, J.E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Characterization of a cytochrome P450 that catalyzes the O-demethylation of lignin-derived benzoates.
J.Biol.Chem., 2024
|
|
8TYO
 
 | |
9B8Q
 
 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3, peripheral stalks | | Descriptor: | V-type proton ATPase 116 kDa subunit a isoform 1, V-type proton ATPase subunit C 1, V-type proton ATPase subunit E 1, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-03-31 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
9B8O
 
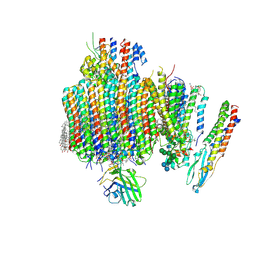 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3, Vo | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-03-31 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
4V4S
 
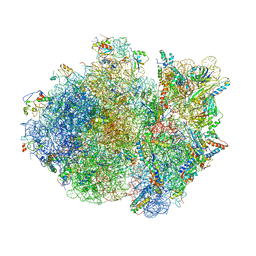 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.76 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
9G9R
 
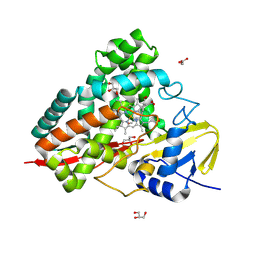 | | Crystal structure of PbdA bound to p-ethylbenzoate | | Descriptor: | 4-ethylbenzoic acid, Cytochrome P450 CYP199, GLYCEROL, ... | | Authors: | Hinchen, D.J, Wolf, M.E, Eltis, L.D, McGeehan, J.E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of a cytochrome P450 that catalyzes the O-demethylation of lignin-derived benzoates.
J.Biol.Chem., 2024
|
|
8TYL
 
 | |
