6QWF
 
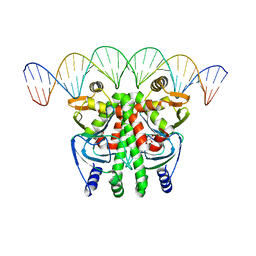 | | The Transcriptional Regulator PrfA-A94V mutant from Listeria Monocytogenes in complex with a 30-bp operator PrfA-box motif | | Descriptor: | DNA (30-MER), GLYCEROL, Listeriolysin positive regulatory factor A | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
7LOE
 
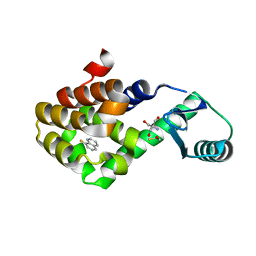 | | T4 lysozyme mutant L99A in complex with 1-fluoranylnaphthalene | | Descriptor: | 1-fluoranylnaphthalene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6T2K
 
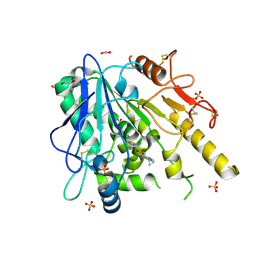 | | Furano[2,3-d]prymidine amides as Notum inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-(6-chloranyl-7-cyclopropyl-thieno[3,2-d]pyrimidin-4-yl)sulfanylethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Scaffold-hopping identifies furano[2,3-d]pyrimidine amides as potent Notum inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7LOA
 
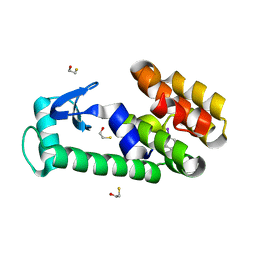 | | T4 lysozyme mutant L99A in complex with 3-fluoroiodobenzene | | Descriptor: | 1-fluoranyl-3-iodanyl-benzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LOF
 
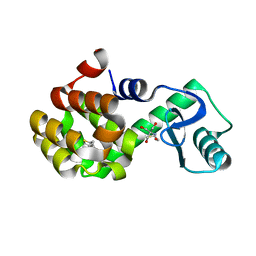 | | T4 lysozyme mutant L99A in complex with 2-butylthiophene | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-butylthiophene, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2ITE
 
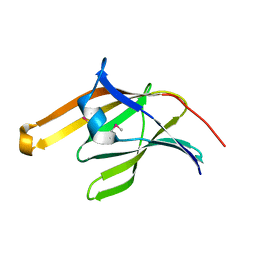 | | Crystal structure of the IsdA NEAT domain from Staphylococcus aureus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Iron-regulated surface determinant protein A | | Authors: | Grigg, J.C, Vermeiren, C.L, Heinrichs, D.E, Murphy, M.E. | | Deposit date: | 2006-10-19 | | Release date: | 2006-12-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Haem recognition by a Staphylococcus aureus NEAT domain.
Mol.Microbiol., 63, 2007
|
|
6QOC
 
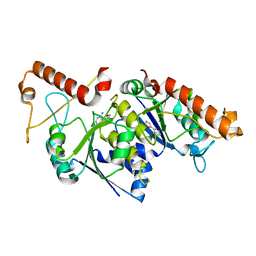 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 6 (2-(1,3-oxazol-5-yl) aniline) | | Descriptor: | 2-(1,3-oxazol-5-yl)aniline, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6QZ8
 
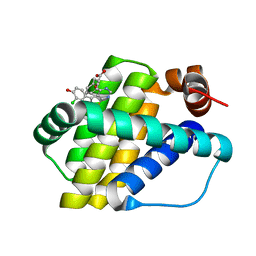 | | Structure of Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QOK
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 14 (Methyl 2-aminopyridine-4-carboxylate) | | Descriptor: | SULFATE ION, methyl 2-azanylpyridine-4-carboxylate, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6QOS
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 22 (Ethyl 1H-pyrazole-4-carboxylate) | | Descriptor: | SULFATE ION, ethyl 1~{H}-pyrazole-4-carboxylate, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
7Q39
 
 | |
4XFS
 
 | | Structure of IL-18 SER Mutant I | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Interleukin-18 | | Authors: | Krumm, B.E, Meng, X, Xiang, Y, Deng, J. | | Deposit date: | 2014-12-28 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystallization of interleukin-18 for structure-based inhibitor design.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6H76
 
 | | SiaP in complex with Neu5Ac (RT) | | Descriptor: | CESIUM ION, CHLORIDE ION, N-acetyl-beta-neuraminic acid, ... | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
6QQY
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 5-azanyl-3-[1-(pyridin-2-ylmethyl)indol-6-yl]-1~{H}-pyrazole-4-carbonitrile, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
5EGU
 
 | | FIRST DOMAIN OF HUMAN BROMODOMAIN BRD4 IN COMPLEX WITH INHIBITOR 3-Butyl-8-(6-butyl-5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-ylsulfanylmethyl)-7-ethyl-3,7-dihydropurine-2,6-dione | | Descriptor: | 1,2-ETHANEDIOL, 3-butyl-8-[(6-butyl-5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)sulfanylmethyl]-7-ethyl-purine-2,6-dione, Bromodomain-containing protein 4 | | Authors: | Raux, B, Rebuffet, E, Betzi, S, Priet, S, Morelli, X. | | Deposit date: | 2015-10-27 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Exploring Selective Inhibition of the First Bromodomain of the Human Bromodomain and Extra-terminal Domain (BET) Proteins.
J.Med.Chem., 59, 2016
|
|
6QR6
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 5-azanyl-3-[1-[[4-(pyrrolidin-1-ylmethyl)phenyl]methyl]indol-6-yl]-1~{H}-pyrazole-4-carbonitrile, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
3I7C
 
 | |
6QWM
 
 | | The Transcriptional Regulator PrfA-A218G mutant from Listeria Monocytogenes in complex with a 30-bp operator PrfA-box motif | | Descriptor: | DNA (30-MER), GLYCEROL, Listeriolysin positive regulatory factor A | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
2IZS
 
 | | Structure of casein kinase gamma 3 in complex with inhibitor | | Descriptor: | CASEIN KINASE I ISOFORM GAMMA-3, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bunkoczi, G, Salah, E, Rellos, P, Das, S, Fedorov, O, Savitsky, P, Debreczeni, J.E, Gileadi, O, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, von Delft, F, Knapp, S. | | Deposit date: | 2006-07-26 | | Release date: | 2006-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibitor Binding by Casein Kinases
To be Published
|
|
5ES3
 
 | | Co-crystal structure of LDH liganded with oxamate | | Descriptor: | L-lactate dehydrogenase A chain, OXAMIC ACID | | Authors: | Nowicki, M.W, Wear, M.A, McNae, I.W, Blackburn, E.A. | | Deposit date: | 2015-11-16 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Streamlined, Automated Protocol for the Production of Milligram Quantities of Untagged Recombinant Rat Lactate Dehydrogenase A Using AKTAxpressTM.
Plos One, 10, 2015
|
|
7TOC
 
 | | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris | | Descriptor: | ACETIC ACID, Ketol-acid reductoisomerase, mitochondrial, ... | | Authors: | Kim, Y, Evdokimova, E, Di, R, Stogios, P, Savchenko, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris
To Be Published
|
|
5EJJ
 
 | | Crystal structure of UfSP from C.elegans | | Descriptor: | Ufm1-specific protease | | Authors: | Kim, K, Ha, B, Kim, E.E. | | Deposit date: | 2015-11-02 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The MPN domain of Caenorhabditis elegans UfSP modulates both substrate recognition and deufmylation activity
Biochem. Biophys. Res. Commun., 476, 2016
|
|
5HMM
 
 | | Crystal Structure of T5 D15 Protein Co-crystallized with Metal Ions | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exodeoxyribonuclease, ... | | Authors: | Flemming, C.S, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-16 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8FHJ
 
 | | Crystal structure of a FAD monooxygenease from Methylocystis sp. Strain SB2 | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Stewart, A.M, Sawaya, M.R, Stewart, C.E. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of MbnF: an NADPH-dependent flavin monooxygenase from Methylocystis strain SB2.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8DD1
 
 | | SARS-CoV-2 Main Protease (Mpro) H164N Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
