7D25
 
 | | Hsp90 alpha N-terminal domain in complex with a 14 compund | | Descriptor: | 6-chloranyl-9-[(4-methylphenyl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
5TMM
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC analog, (E)-6-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)hex-5-enoic acid | | Descriptor: | 6-{4-[(1S,4S,6S)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}hex-5-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
6I4J
 
 | | Crystal Structure of Plasmodium falciparum actin I (F54Y mutant) in the Mg-ADP state | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin-1, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
6I4L
 
 | | Crystal Structure of Plasmodium falciparum actin I (G115A mutant) in the Mg-K-ATP/ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-1, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
6U9M
 
 | | MLL1 SET N3861I/Q3867L bound to inhibitor 16 (TC-5109) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(thiophen-2-yl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
7H2C
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z1328968520 | | Descriptor: | 4-[(2-methylsulfonylimidazol-1-yl)methyl]-1,3-thiazole, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
7H2O
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z234898257 | | Descriptor: | DIMETHYL SULFOXIDE, N-methyl-1-([1,2,4]triazolo[4,3-a]pyridin-3-yl)methanamine, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
7H0X
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0013255-001 | | Descriptor: | (4M)-1-methyl-4-(4-{[(1S)-2-methyl-1-(3-oxo-3,4-dihydro-2H-1,4-benzoxazin-6-yl)propyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-1H-pyrazole-5-carbonitrile, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
6I7S
 
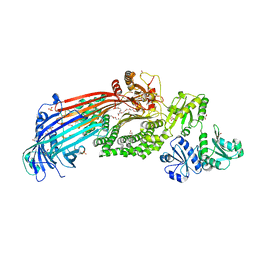 | | Microsomal triglyceride transfer protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Biterova, E, Isupov, M.N, Keegan, R.M, Lebedev, A.A, Ruddock, L.W. | | Deposit date: | 2018-11-17 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of human microsomal triglyceride transfer protein.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7H2E
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z1428159350 | | Descriptor: | (1S)-1-(1-phenyl-1H-1,2,3-triazol-4-yl)ethan-1-amine, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
6I3B
 
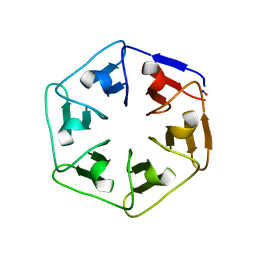 | |
3SX9
 
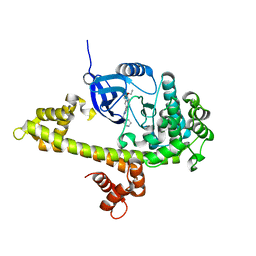 | |
2E9T
 
 | | Foot-and-mouth disease virus RNA-polymerase RNA dependent in complex with a template-primer RNA and 5F-UTP | | Descriptor: | 5'-R(*GP*GP*GP*CP*CP*CP*(5FU))-3', 5'-R(P*UP*AP*GP*GP*GP*CP*CP*C)-3', MAGNESIUM ION, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequential structures provide insights into the fidelity of RNA replication
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7H2L
 
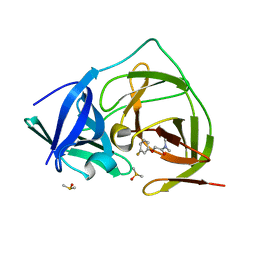 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z19755216 | | Descriptor: | 2-(3-fluorophenoxy)-N,N-dimethylacetamide, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.366 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
1B4D
 
 | | AMIDOCARBAMATE INHIBITOR OF GLYCOGEN PHOSPHORYLASE | | Descriptor: | 1-DEOXY-1-METHOXYCARBAMIDO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, INOSINIC ACID, PROTEIN (GLYCOGEN PHOSPHORYLASE B), ... | | Authors: | Tsitsanou, K.E, Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Gregoriou, M, Watson, K.A, Johnson, L.N, Fleet, G.W.J. | | Deposit date: | 1998-12-18 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of commonly used cryoprotectants on glycogen phosphorylase activity and structure.
Protein Sci., 8, 1999
|
|
6I4F
 
 | | Crystal Structure of Plasmodium falciparum actin I (A272W mutant) in the Mg-K-ATP/ADP state | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
4OO1
 
 | |
2R0I
 
 | |
6ICJ
 
 | | Crystal structure of PPARgamma with compound BR102375K | | Descriptor: | 2-butyl-5-[(3-tert-butyl-1,2,4-oxadiazol-5-yl)methyl]-6-methyl-3-{[2'-(5-oxo-4,5-dihydro-1,2,4-oxadiazol-3-yl)[1,1'-biphenyl]-4-yl]methyl}pyrimidin-4(3H)-one, GLYCEROL, Nuclear receptor coactivator 1, ... | | Authors: | Hong, E, Chin, J, Jang, T.H, Kim, K.H, Jung, W, Kim, S.H. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Crystal structure of PPARgamma with compound BR102375K
To Be Published
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6OFZ
 
 | | Crystal structure of human WDR5 | | Descriptor: | WD repeat-containing protein 5 | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
1N18
 
 | | Thermostable mutant of Human Superoxide Dismutase, C6A, C111S | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Cardoso, R.M.F, Thayer, M.M, DiDonato, M, Lo, T.P, Bruns, C.K, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2002-10-16 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into Lou Gehrig's disease from the structure and instability of the A4V mutant of human Cu,Zn superoxide dismutase.
J.Mol.Biol., 324, 2002
|
|
1N0J
 
 | |
1N0T
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with the antagonist (S)-ATPO at 2.1 A resolution. | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Greenwood, J.R, Liljefors, T, Lunn, M.-L, Egebjerg, J, Larsen, I.K, Gouaux, E, Kastrup, J.S. | | Deposit date: | 2002-10-15 | | Release date: | 2003-03-04 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive antagonism of AMPA receptors by ligands of
different classes: crystal structure of ATPO bound to the
GluR2 ligand-binding core, in comparison with DNQX.
J.Med.Chem., 46, 2003
|
|
4G1Q
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with Rilpivirine (TMC278, Edurant), a non-nucleoside rt-inhibiting drug | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, MAGNESIUM ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2012-07-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Snapshot of the equilibrium dynamics of a drug bound to HIV-1 reverse transcriptase.
Nat Chem, 5, 2013
|
|
