6U2M
 
 | | Crystal structure of a HaloTag-based calcium indicator, HaloCaMP V2, bound to JF635 | | Descriptor: | (1E,3S)-1-{10-[2-carboxy-5-({2-[2-(hexyloxy)ethoxy]ethyl}carbamoyl)phenyl]-7-(3-fluoroazetidin-1-yl)-5,5-dimethyldibenz o[b,e]silin-3(5H)-ylidene}-3-fluoroazetidin-1-ium, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Deo, C, Schreiter, E.R. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The HaloTag as a general scaffold for far-red tunable chemigenetic indicators.
Nat.Chem.Biol., 17, 2021
|
|
5LLA
 
 | | Crystal structure of human carbonic anhydrase isozyme XIII with 4-(1H-benzimidazol-1-ylacetyl)-2-chlorobenzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(benzimidazol-1-yl)ethanoyl]-2-chloranyl-benzenesulfonamide, CITRIC ACID, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2016-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combinatorial Design of Isoform-Selective N-Alkylated Benzimidazole-Based Inhibitors of Carbonic Anhydrases
Chemistryselect, 2017
|
|
5IJ0
 
 | | Cryo EM density of microtubule assembled from human TUBB3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ti, S.-C, Pamula, M.C, Howes, S.C, Duellberg, C, Nicholas, C.I, Kleiner, R.E, Forth, S, Surrey, T, Nogales, E, Kapoor, T.M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mutations in Human Tubulin Proximal to the Kinesin-Binding Site Alter Dynamic Instability at Microtubule Plus- and Minus-Ends.
Dev.Cell, 37, 2016
|
|
5LOE
 
 | | Structure of full length Cody from Bacillus subtilis in complex with Ile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, ISOLEUCINE | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2016-08-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Branched-chain Amino Acid and GTP-sensing Global Regulator, CodY, from Bacillus subtilis.
J. Biol. Chem., 292, 2017
|
|
6O6C
 
 | | RNA polymerase II elongation complex arrested at a CPD lesion | | Descriptor: | DNA (27-MER), DNA (5'-D(P*GP*GP*AP*GP*AP*AP*GP*GP*AP*GP*CP*AP*GP*AP*GP*C)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Lahiri, I, Leshziner, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | 3.1 angstrom structure of yeast RNA polymerase II elongation complex stalled at a cyclobutane pyrimidine dimer lesion solved using streptavidin affinity grids.
J.Struct.Biol., 207, 2019
|
|
5LQY
 
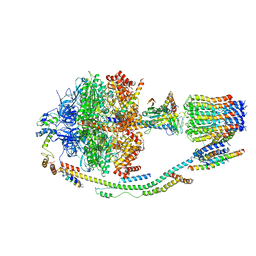 | | Structure of F-ATPase from Pichia angusta, in state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase OSCP subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
8DMJ
 
 | |
6OBL
 
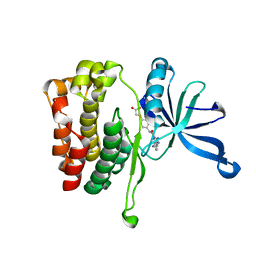 | | JAK2 JH2 in complex with JAK168 | | Descriptor: | Tyrosine-protein kinase JAK2, [4-({5-amino-3-[(4-cyanophenyl)amino]-1H-1,2,4-triazole-1-carbonyl}amino)phenoxy]acetic acid | | Authors: | Krimmer, S.G, Liosi, M.E, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2019-03-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core.
J.Med.Chem., 63, 2020
|
|
8DPM
 
 | |
8DPL
 
 | |
6TV3
 
 | | HumRadA1 in complex with 3-amino-2-naphthoic acid | | Descriptor: | 3-azanylnaphthalene-2-carboxylic acid, DNA repair and recombination protein RadA, GLYCEROL, ... | | Authors: | Marsh, M.E, Hyvonen, M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
5LL5
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 4-(1H-benzimidazol-1-ylacetyl)benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(benzimidazol-1-yl)ethanoyl]benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2016-07-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Combinatorial Design of Isoform-Selective N-Alkylated Benzimidazole-Based Inhibitors of Carbonic Anhydrases
Chemistryselect, 2017
|
|
6OGG
 
 | | 70S termination complex with RF2 bound to the UGA codon. Rotated ribosome with RF2 bound (Structure IV). | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-02 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6OI3
 
 | | Crystal structure of human WDR5 in complex with monomethyl H3R2 peptide | | Descriptor: | GLYCEROL, Monomethyl H3R2 peptide, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6ORO
 
 | | Modified BG505 SOSIP-based immunogen RC1 in complex with the elicited V3-glycan patch antibody Ab874NHP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab874NHP antibody Fab heavy chain, ... | | Authors: | Abernathy, M.E, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques.
Nature, 570, 2019
|
|
6OSI
 
 | | Unmodified tRNA(Pro) bound to Thermus thermophilus 70S (near cognate) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Subaramanian, S, Hong, S, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2019-05-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4.14 Å) | | Cite: | Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.
Elife, 9, 2020
|
|
5LXS
 
 | | Tubulin-KS-1-199-32 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Steinmetz, M.O. | | Deposit date: | 2016-09-22 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Microtubule Stabilization by Discodermolide.
Chembiochem, 18, 2017
|
|
5LQX
 
 | | Structure of F-ATPase from Pichia angusta, state3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase AAP1 subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5LSM
 
 | |
8OID
 
 | | Cryo-EM structure of ADP-bound, filamentous beta-actin harboring the N111S mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Blanc, F.E.C, Roy, A, Belyy, A, Hofnagel, O, Hummer, G, Bieling, P, Raunser, S. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Molecular mechanisms of inorganic-phosphate release from the core and barbed end of actin filaments.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6UJX
 
 | | HIV-1 wild-type reverse transcriptase-DNA complex with (-)-FTC-TP | | Descriptor: | DNA primer, DNA template, MAGNESIUM ION, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2019-10-03 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.70451212 Å) | | Cite: | Elucidating molecular interactions ofL-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance.
Commun Biol, 2, 2019
|
|
4XIW
 
 | | Carbonic anhydrase Cah3 from Chlamydomonas reinhardtii in complex with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase, alpha type, ... | | Authors: | Hainzl, T, Grundstrom, C, Benlloch, R, Shevela, D, Shutova, T, Messinger, J, Samuelsson, G, Sauer-Eriksson, A.E. | | Deposit date: | 2015-01-07 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Functional Characterization of Photosystem II-Associated Carbonic Anhydrase CAH3 in Chlamydomonas reinhardtii.
Plant Physiol., 167, 2015
|
|
5M4V
 
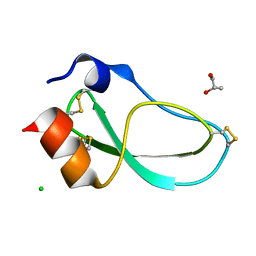 | | X-ray structure of the mambaquaretin-1, a selective antagonist of the vasopressin type 2 receptor | | Descriptor: | CHLORIDE ION, Mambaquaretin-1, S-1,2-PROPANEDIOL | | Authors: | Stura, E.A, Vera, L, Ciolek, J, Mourier, G, Gilles, N. | | Deposit date: | 2016-10-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Green mamba peptide targets type-2 vasopressin receptor against polycystic kidney disease.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8ONL
 
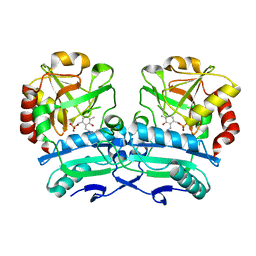 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONJ
 
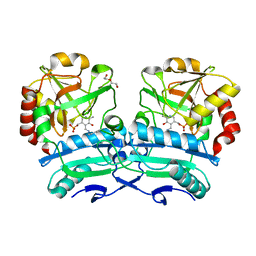 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant R88L | | Descriptor: | Aminotransferase class IV, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
