5HOC
 
 | | p73 homo-tetramerization domain mutant II | | 分子名称: | Tumor protein p73 | | 著者 | Coutandin, D, Krojer, T, Salah, E, Mathea, S, Sumyk, M, Knapp, S, Dotsch, V. | | 登録日 | 2016-01-19 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.36007786 Å) | | 主引用文献 | Mechanism of TAp73 inhibition by Delta Np63 and structural basis of p63/p73 hetero-tetramerization.
Cell Death Differ., 23, 2016
|
|
5HOU
 
 | | Solution Structure of p53TAD-TAZ1 | | 分子名称: | Cellular tumor antigen p53,CREB-binding protein fusion protein, ZINC ION | | 著者 | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | 登録日 | 2016-01-19 | | 公開日 | 2016-03-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4W9X
 
 | | Crystal Structure of BMP-2-inducible kinase in complex with baricitinib | | 分子名称: | 1,2-ETHANEDIOL, BMP-2-inducible protein kinase, Baricitinib | | 著者 | Sorrell, F.J, Elkins, J.M, Krojer, T, Williams, E, Savitsky, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2014-08-28 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Family-wide Structural Analysis of Human Numb-Associated Protein Kinases.
Structure, 24, 2016
|
|
4EKA
 
 | | Final Thaumatin Structure for Radiation Damage Experiment at 25 K | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Warkentin, M, Badeau, R, Hopkins, J.B, Thorne, R.E. | | 登録日 | 2012-04-09 | | 公開日 | 2012-08-29 | | 最終更新日 | 2013-01-23 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Spatial distribution of radiation damage to crystalline proteins at 25-300 K.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5HCK
 
 | | HUMAN HCK SH3 DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | HEMATOPOIETIC CELL KINASE | | 著者 | Horita, D.A, Baldisseri, D.M, Zhang, W, Altieri, A.S, Smithgall, T.E, Gmeiner, W.H, Byrd, R.A. | | 登録日 | 1998-03-09 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the human Hck SH3 domain and identification of its ligand binding site.
J.Mol.Biol., 278, 1998
|
|
4EQN
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/I72K at cryogenic temperature | | 分子名称: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | 著者 | Robinson, A.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | 登録日 | 2012-04-19 | | 公開日 | 2012-05-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/I72K at cryogenic temperature
To be Published
|
|
5I4C
 
 | | Crystal structure of non-phosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli | | 分子名称: | Transcriptional regulatory protein RcsB | | 著者 | Filippova, E.V, Wawrzak, Z, Minasov, G, Ruan, J, Pshenychnyi, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-02-11 | | 公開日 | 2016-10-12 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of nonphosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli.
Protein Sci., 25, 2016
|
|
4EVO
 
 | |
4EEP
 
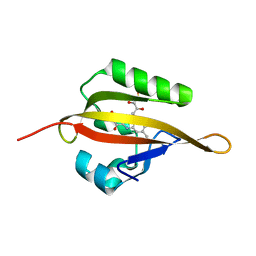 | | Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 | | 分子名称: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | 著者 | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2012-03-28 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4W8R
 
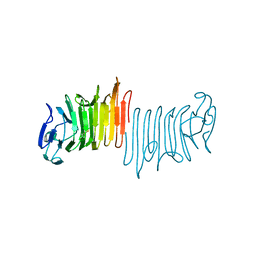 | |
4W8X
 
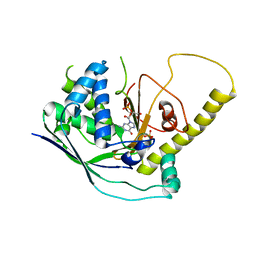 | | Crystal Structure of Cmr1 from Pyrococcus furiosus bound to a nucleotide | | 分子名称: | CRISPR system Cmr subunit Cmr1-1, GUANOSINE-3'-MONOPHOSPHATE, PHOSPHATE ION | | 著者 | Benda, C, Ebert, J, Baumgaertner, M, Conti, E. | | 登録日 | 2014-08-26 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Model of a CRISPR RNA-Silencing Complex Reveals the RNA-Target Cleavage Activity in Cmr4.
Mol.Cell, 56, 2014
|
|
5I6Y
 
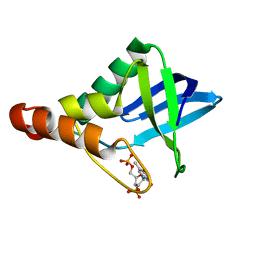 | |
4EK0
 
 | | Initial Thaumatin Structure for Radiation Damage Experiment at 25 K | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Warkentin, M, Badeau, R, Hopkins, J.B, Thorne, R.E. | | 登録日 | 2012-04-08 | | 公開日 | 2012-08-29 | | 最終更新日 | 2013-01-23 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Spatial distribution of radiation damage to crystalline proteins at 25-300 K.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4EKH
 
 | | Final Thaumatin Structure for Radiation Damage Experiment at 100 K | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Warkentin, M, Badeau, R, Hopkins, J.B, Thorne, R.E. | | 登録日 | 2012-04-09 | | 公開日 | 2012-08-29 | | 最終更新日 | 2013-01-23 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Spatial distribution of radiation damage to crystalline proteins at 25-300 K.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5I9S
 
 | | MicroED structure of proteinase K at 1.75 A resolution | | 分子名称: | Proteinase K, SULFATE ION | | 著者 | Hattne, J, Shi, D, de la Cruz, M.J, Reyes, F.E, Gonen, T. | | 登録日 | 2016-02-20 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.75 Å) | | 主引用文献 | Modeling truncated pixel values of faint reflections in MicroED images.
J.Appl.Crystallogr., 49, 2016
|
|
4ELA
 
 | | Final Thaumatin Structure for Radiation Damage Experiment at 300 K | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Warkentin, M, Badeau, R, Hopkins, J.B, Thorne, R.E. | | 登録日 | 2012-04-10 | | 公開日 | 2012-08-29 | | 最終更新日 | 2013-01-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Spatial distribution of radiation damage to crystalline proteins at 25-300 K.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4TYV
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose | | 分子名称: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose | | 著者 | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | 登録日 | 2014-07-09 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
5I45
 
 | | 1.35 Angstrom Crystal Structure of C-terminal Domain of Glycosyl Transferase Group 1 Family Protein (LpcC) from Francisella tularensis. | | 分子名称: | Glycosyl transferases group 1 family protein | | 著者 | Minasov, G, Filippova, E, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-02-11 | | 公開日 | 2016-02-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | 1.35 Angstrom Crystal Structure of C-terminal Domain of Glycosyl Transferase Group 1 Family Protein (LpcC) from Francisella tularensis.
To Be Published
|
|
6EBE
 
 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | 分子名称: | 4-hydroxy-3-nitro-5-({[4-(trifluoromethyl)phenyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | 著者 | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | 登録日 | 2018-08-06 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
4TUH
 
 | | Bcl-xL in complex with inhibitor (Compound 10) | | 分子名称: | 1,2-ETHANEDIOL, 2-[8-(1,3-benzothiazol-2-ylcarbamoyl)-3,4-dihydroisoquinolin-2(1H)-yl]-5-{3-[4-(1H-pyrazolo[3,4-d]pyrimidin-1-yl)phenoxy]propyl}-1,3-thiazole-4-carboxylic acid, ACETATE ION, ... | | 著者 | Czabotar, P.E, Lessense, G, Smith, B.J, Colman, P.M. | | 登録日 | 2014-06-24 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Guided Rescaffolding of Selective Antagonists of BCL-XL.
Acs Med.Chem.Lett., 5, 2014
|
|
4U3G
 
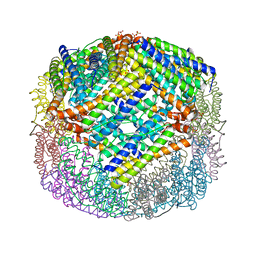 | | Crystal structure of Escherichia coli bacterioferritin mutant D132F | | 分子名称: | Bacterioferritin, SULFATE ION | | 著者 | Wong, S.G, Grigg, J.C, Le Brun, N.E, Moore, G.R, Murphy, M.E.P, Mauk, A.G. | | 登録日 | 2014-07-21 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The B-type Channel Is a Major Route for Iron Entry into the Ferroxidase Center and Central Cavity of Bacterioferritin.
J.Biol.Chem., 290, 2015
|
|
4UD7
 
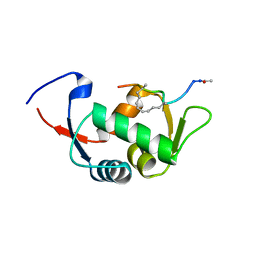 | | Structure of the stapled peptide YS-02 bound to MDM2 | | 分子名称: | MDM2, YS-02 | | 著者 | Tan, Y.S, Reeks, J, Brown, C.J, Jennings, C.E, Eapen, R.S, Tng, Q.S, Thean, D, Ying, Y.T, Gago, F.J.F, Lane, D.P, Noble, M.E.M, Verma, C. | | 登録日 | 2014-12-08 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Benzene Probes in Molecular Dynamics Simulations Reveal Novel Binding Sites for Ligand Design.
J Phys Chem Lett, 7, 2016
|
|
4UE1
 
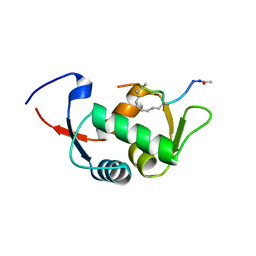 | | Structure of the stapled peptide YS-01 bound to MDM2 | | 分子名称: | E3 UBIQUITIN-PROTEIN LIGASE MDM2, YS-01 | | 著者 | Tan, Y.S, Reeks, J, Brown, C.J, Jennings, C.E, Eapen, R.S, Tng, Q.S, Thean, D, Ying, Y.T, Gago, F.J.F, Lane, D.P, Noble, M.E.M, Verma, C. | | 登録日 | 2014-12-14 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Benzene Probes in Molecular Dynamics Simulations Reveal Novel Binding Sites for Ligand Design.
J Phys Chem Lett, 7, 2016
|
|
4USE
 
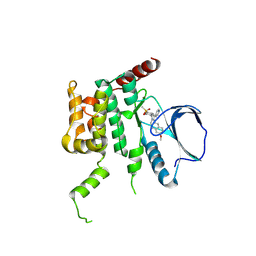 | | Human STK10 (LOK) with SB-633825 | | 分子名称: | 4-{5-(6-methoxynaphthalen-2-yl)-1-methyl-2-[2-methyl-4-(methylsulfonyl)phenyl]-1H-imidazol-4-yl}pyridine, SERINE/THREONINE-PROTEIN KINASE 10 | | 著者 | Elkins, J.M, Salah, E, Szklarz, M, von Delft, F, Canning, P, Raynor, J, Bountra, C, Edwards, A.M, Knapp, S. | | 登録日 | 2014-07-07 | | 公開日 | 2015-07-22 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Comprehensive Characterization of the Published Kinase Inhibitor Set.
Nat.Biotechnol., 34, 2016
|
|
1GJE
 
 | | Peptide Antagonist of IGFBP-1, Minimized Average Structure | | 分子名称: | IGFBP-1 antagonist | | 著者 | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | 登録日 | 2001-05-11 | | 公開日 | 2001-05-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
