6BW5
 
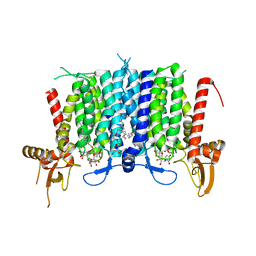 | | Human GPT (DPAGT1) in complex with tunicamycin | | 分子名称: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | 著者 | Yoo, J, Kuk, A.C.Y, Mashalidis, E.H, Lee, S.-Y. | | 登録日 | 2017-12-14 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7UVK
 
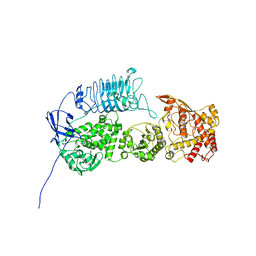 | | G. haemolysans IgA1 protease | | 分子名称: | IgA1 Protease | | 著者 | Eisenmesser, E.Z, Zheng, H. | | 登録日 | 2022-05-02 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | A substrate-induced gating mechanism is conserved among Gram-positive IgA1 metalloproteases.
Commun Biol, 5, 2022
|
|
2NV7
 
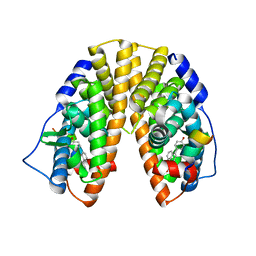 | | Crystal Structure of Estrogen Receptor Beta Complexed with WAY-555 | | 分子名称: | 4-(4-HYDROXYPHENYL)-1-NAPHTHALDEHYDE OXIME, Estrogen receptor beta, Nuclear receptor coactivator 1 | | 著者 | Mewshaw, R.E, Bowen, M.S, Harris, H.A, Xu, Z.B, Manas, E.S, Cohn, S.T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-11-10 | | 公開日 | 2007-08-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | ERbeta ligands. Part 5: synthesis and structure-activity relationships of a series of 4'-hydroxyphenyl-aryl-carbaldehyde oxime derivatives.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4GWA
 
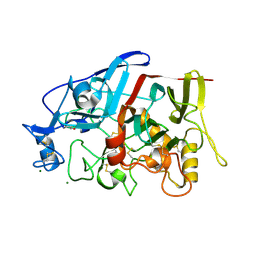 | | Crystal Structure of a GH7 Family Cellobiohydrolase from Limnoria quadripunctata | | 分子名称: | GH7 family protein, MAGNESIUM ION | | 著者 | McGeehan, J.E, Martin, R.N.A, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | 登録日 | 2012-09-01 | | 公開日 | 2013-06-12 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6OU1
 
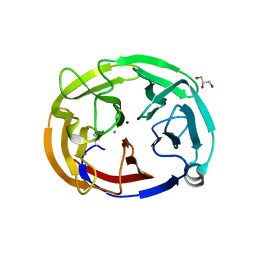 | | Crystal Structure of the Computationally-derived 21-Variant of the Myocilin Olfactomedin Domain | | 分子名称: | CALCIUM ION, GLYCEROL, Myocilin, ... | | 著者 | Hill, S.E, Kwon, M.S, Lieberman, R.L. | | 登録日 | 2019-05-03 | | 公開日 | 2019-07-03 | | 最終更新日 | 2020-01-15 | | 実験手法 | X-RAY DIFFRACTION (1.878 Å) | | 主引用文献 | Stable calcium-free myocilin olfactomedin domain variants reveal challenges in differentiating between benign and glaucoma-causing mutations.
J.Biol.Chem., 294, 2019
|
|
5IXH
 
 | | Crystal Structure of Burkholderia cenocepacia BcnA | | 分子名称: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, ... | | 著者 | Loutet, S.A, Murphy, M.E.P. | | 登録日 | 2016-03-23 | | 公開日 | 2017-03-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Antibiotic Capture by Bacterial Lipocalins Uncovers an Extracellular Mechanism of Intrinsic Antibiotic Resistance.
MBio, 8, 2017
|
|
8CEX
 
 | |
5D2C
 
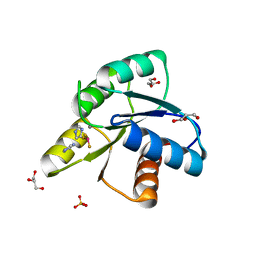 | | Reaction of phosphorylated CheY with imidazole 1 of 3 | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | 著者 | Page, S, Silversmith, R.E, Bourret, R.B, Collins, E.J. | | 登録日 | 2015-08-05 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Imidazole as a Small Molecule Analogue in Two-Component Signal Transduction.
Biochemistry, 54, 2015
|
|
6C23
 
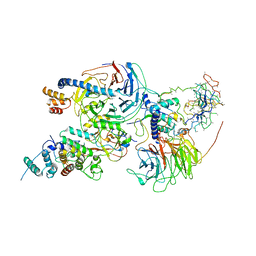 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State | | 分子名称: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | 著者 | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | 登録日 | 2018-01-05 | | 公開日 | 2018-01-24 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
6YOB
 
 | | Structure of Lysozyme from COC IMISX setup collected by rotation serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, ACETIC ACID, BROMIDE ION, ... | | 著者 | Huang, C.-Y, Martiel, I, Villanueva-Perez, P, Panepucci, E, Caffrey, M, Wang, M. | | 登録日 | 2020-04-14 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Iucrj, 7, 2020
|
|
6FVU
 
 | | 26S proteasome, s2 state | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | 著者 | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | 登録日 | 2018-03-05 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6YOG
 
 | | Structure of PepTSt from COC IMISX setup collected by still serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | | 分子名称: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 2-(2-METHOXYETHOXY)ETHANOL, Di-or tripeptide:H+ symporter, ... | | 著者 | Huang, C.-Y, Martiel, I, Villanueva-Perez, P, Panepucci, E, Caffrey, M, Wang, M. | | 登録日 | 2020-04-14 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Iucrj, 7, 2020
|
|
6OWV
 
 | | Crystal structure of a Human Cardiac Calsequestrin Filament | | 分子名称: | CHLORIDE ION, Calsequestrin-2, SULFATE ION | | 著者 | Titus, E.W, Deiter, F.H, Shi, C, Jura, N, Deo, R.C. | | 登録日 | 2019-05-12 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | The structure of a calsequestrin filament reveals mechanisms of familial arrhythmia.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2VZ5
 
 | | Structure of the PDZ domain of Tax1 (human T-cell leukemia virus type I) binding protein 3 | | 分子名称: | CHLORIDE ION, IMIDAZOLE, TAX1-BINDING PROTEIN 3, ... | | 著者 | Murray, J.W, Shafqat, N, Yue, W, Pilka, E, Johannsson, C, Salah, E, Cooper, C, Elkins, J.M, Pike, A.C, Roos, A, Filippakopoulos, P, von Delft, F, Wickstroem, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Oppermann, U. | | 登録日 | 2008-07-30 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.738 Å) | | 主引用文献 | The Structure of the Pdz Domain of Tax1BP
To be Published
|
|
8JXS
 
 | | Structure of nanobody-bound DRD1_PF-6142 complex | | 分子名称: | 4-[3-methyl-4-(6-methylimidazo[1,2-a]pyrazin-5-yl)phenoxy]furo[3,2-c]pyridine, D(1A) dopamine receptor, Fab 8D3 heavy chain, ... | | 著者 | Zhuang, Y, Xu, Y, Fan, L, Wang, S, Xu, H.E. | | 登録日 | 2023-07-01 | | 公開日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of psychedelic LSD recognition at dopamine D 1 receptor.
Neuron, 2024
|
|
1JZK
 
 | | Crystal Structure of Scapharca inaequivalvis HbI, I114F mutant (deoxy) | | 分子名称: | GLOBIN I - ARK SHELL, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Knapp, J.E, Gibson, Q.H, Cushing, L, Royer Jr, W.E. | | 登録日 | 2001-09-16 | | 公開日 | 2001-12-19 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Restricting the Ligand-Linked Heme Movement in Scapharca Dimeric Hemoglobin Reveals Tight Coupling between Distal and Proximal
Contributions to Cooperativity.
Biochemistry, 40, 2001
|
|
6YDF
 
 | | X-ray structure of LPMO. | | 分子名称: | COPPER (II) ION, LPMO lytic polysaccharide monooxygenase, SULFATE ION | | 著者 | Tandrup, T, Tryfona, T, Frandsen, K.E.H, Johansen, K.S, Dupree, P, Lo Leggio, L. | | 登録日 | 2020-03-20 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Oligosaccharide Binding and Thermostability of Two Related AA9 Lytic Polysaccharide Monooxygenases.
Biochemistry, 59, 2020
|
|
8CEY
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH11233 | | 分子名称: | N-glycosylase/DNA lyase, NICKEL (II) ION, ~{N}-(3,3-dimethylcyclobutyl)imidazo[5,1-b][1,3]thiazole-3-carboxamide | | 著者 | Kosenina, S, Scaletti, E.R, Stenmark, P. | | 登録日 | 2023-02-02 | | 公開日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH11233
To Be Published
|
|
2NVA
 
 | | The X-ray crystal structure of the Paramecium bursaria Chlorella virus arginine decarboxylase bound to agmatine | | 分子名称: | (4-{[(4-{[AMINO(IMINO)METHYL]AMINO}BUTYL)AMINO]METHYL}-5-HYDROXY-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, arginine decarboxylase, A207R protein | | 著者 | Shah, R.H, Akella, R, Goldsmith, E, Phillips, M.A. | | 登録日 | 2006-11-11 | | 公開日 | 2007-03-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray Structure of Paramecium bursaria Chlorella Virus Arginine Decarboxylase: Insight into the Structural Basis for Substrate Specificity.
Biochemistry, 46, 2007
|
|
1GIA
 
 | |
6ZHB
 
 | | 3D electron diffraction structure of bovine insulin | | 分子名称: | Insulin, ZINC ION | | 著者 | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Bacia-Verloop, M, Zander, U, McCarthy, A.A, Schoehn, G, Ling, W.L, Abrahams, J.P. | | 登録日 | 2020-06-22 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (3.25 Å) | | 主引用文献 | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6OU3
 
 | |
4F6X
 
 | | Crystal structure of dehydrosqualene synthase (crtm) from s. aureus complexed with bph-1112 | | 分子名称: | 1-[3-(hexyloxy)benzyl]-4-hydroxy-2-oxo-1,2-dihydropyridine-3-carboxylic acid, D(-)-TARTARIC ACID, Dehydrosqualene synthase, ... | | 著者 | Lin, F.-Y, Zhang, Y, Liu, Y.-L, Oldfield, E. | | 登録日 | 2012-05-15 | | 公開日 | 2012-06-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
8JXR
 
 | | Structure of nanobody-bound DRD1_LSD complex | | 分子名称: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, D(1A) dopamine receptor, Fab 8D3 heavy chain, ... | | 著者 | Zhuang, Y, Xu, Y, Fan, L, Wang, S, Xu, H.E. | | 登録日 | 2023-07-01 | | 公開日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (3.57 Å) | | 主引用文献 | Structural basis of psychedelic LSD recognition at dopamine D 1 receptor.
Neuron, 2024
|
|
1J9K
 
 | | CRYSTAL STRUCTURE OF SURE PROTEIN FROM T.MARITIMA IN COMPLEX WITH TUNGSTATE | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, ... | | 著者 | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | 登録日 | 2001-05-27 | | 公開日 | 2001-09-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
