6XUR
 
 | | RNA dodecamer with a 6-hydrazino-2-aminopurine modified base | | 分子名称: | MAGNESIUM ION, RNA dodecamer with a 6-hydrazino-2-aminopurine modified base | | 著者 | Ennifar, E, Micura, R, Gasser, C, Brillet, K. | | 登録日 | 2020-01-21 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Thioguanosine Conversion Enables mRNA-Lifetime Evaluation by RNA Sequencing Using Double Metabolic Labeling (TUC-seq DUAL).
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5NFW
 
 | | Neutron structure of human transthyretin (TTR) S52P mutant at room temperature to 1.8A resolution (quasi-Laue) | | 分子名称: | Transthyretin | | 著者 | Yee, A.W, Moulin, M, Blakeley, M.P, Cooper, J.B, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | 登録日 | 2017-03-16 | | 公開日 | 2019-01-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | 主引用文献 | A molecular mechanism for transthyretin amyloidogenesis.
Nat Commun, 10, 2019
|
|
6FDC
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-32a | | 分子名称: | (2~{R})-1-[3-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, (2~{S})-1-[5-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, 1,2-ETHANEDIOL, ... | | 著者 | Prosdocimi, T, Donini, S, Parisini, E. | | 登録日 | 2017-12-22 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
3MK7
 
 | | The structure of CBB3 cytochrome oxidase | | 分子名称: | 30-mer peptide, CALCIUM ION, COPPER (II) ION, ... | | 著者 | Buschmann, S, Warkentin, E, Michel, H, Ermler, U. | | 登録日 | 2010-04-14 | | 公開日 | 2010-08-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The Structure of cbb3 Cytochrome Oxidase Provides Insights into Proton Pumping
Science, 329, 2010
|
|
3M3K
 
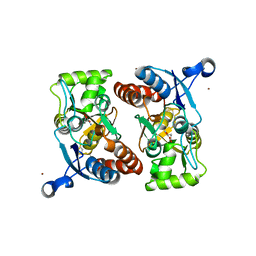 | | Ligand binding domain (S1S2) of GluA3 (flop) | | 分子名称: | GLUTAMIC ACID, Glutamate receptor 3, ZINC ION | | 著者 | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | 登録日 | 2010-03-09 | | 公開日 | 2010-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.793 Å) | | 主引用文献 | Molecular mechanism of flop selectivity and subsite recognition for an AMPA receptor allosteric modulator: structures of GluA2 and GluA3 in complexes with PEPA.
Biochemistry, 49, 2010
|
|
2W9L
 
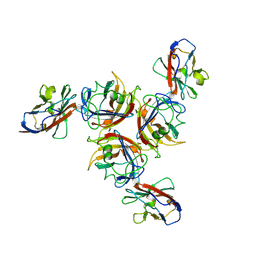 | | CANINE ADENOVIRUS TYPE 2 FIBRE HEAD IN COMPLEX WITH CAR DOMAIN D1 AND SIALIC ACID | | 分子名称: | COXSACKIEVIRUS AND ADENOVIRUS RECEPTOR, FIBRE PROTEIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | 著者 | Seiradake, E, Henaff, D, Wodrich, H, Billet, O, Perreau, M, Hippert, C, Mennechet, F, Schoehn, G, Lortat-Jacob, H, Dreja, H, Ibanes, S, Kalatzis, V, Wang, J.P, Finberg, R.W, Cusack, S, Kremer, E.J. | | 登録日 | 2009-01-26 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | The cell adhesion molecule "CAR" and sialic acid on human erythrocytes influence adenovirus in vivo biodistribution.
PLoS Pathog., 5, 2009
|
|
4ABT
 
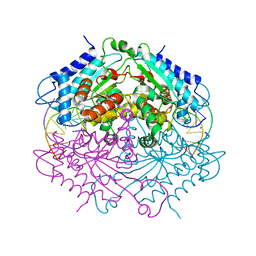 | | Crystal structure of Type IIF restriction endonuclease NgoMIV with cognate uncleaved DNA | | 分子名称: | 5'-D(*TP*GP*CP*GP*CP*CP*GP*GP*CP*GP*CP)-3', CALCIUM ION, TYPE-2 RESTRICTION ENZYME NGOMIV | | 著者 | Manakova, E.N, Grazulis, S, Zaremba, M, Tamulaitiene, G, Golovenko, D, Siksnys, V. | | 登録日 | 2011-12-11 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Structure of Type Iif Restriction Endonuclease Ngomiv with Cognate Uncleaved DNA
To be Published
|
|
6FLM
 
 | |
3MAC
 
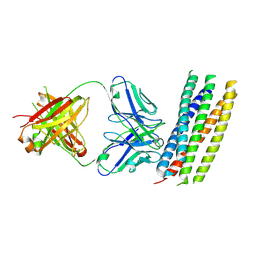 | | crystal structure of GP41-derived protein complexed with fab 8062 | | 分子名称: | Fab8062, Transmembrane glycoprotein | | 著者 | Li, M, Gustchina, E, Louis, J, Gustchina, A, Wlodawer, A, Clore, M. | | 登録日 | 2010-03-23 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis of HIV-1 Neutralization by Affinity Matured Fabs Directed against the Internal Trimeric Coiled-Coil of gp41.
Plos Pathog., 6, 2010
|
|
5F9A
 
 | | Blood group antigen binding adhesin BabA of Helicobacter pylori strain P436 in complex with blood group H Lewis b hexasaccharide | | 分子名称: | Adhesin binding fucosylated histo-blood group antigen,Adhesin,Adhesin binding fucosylated histo-blood group antigen, Nanobody Nb-ER19, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Moonens, K, Gideonsson, P, Subedi, S, Romao, E, Oscarson, S, Muyldermans, S, Boren, T, Remaut, H. | | 登録日 | 2015-12-09 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori.
Cell Host Microbe, 19, 2016
|
|
2OK7
 
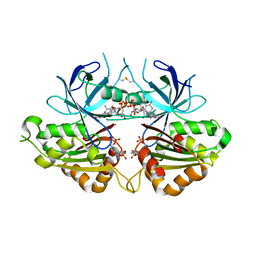 | | Ferredoxin-NADP+ reductase from Plasmodium falciparum with 2'P-AMP | | 分子名称: | ADENOSINE-2'-5'-DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Putative ferredoxin--NADP reductase, ... | | 著者 | Milani, M, Mastrangelo, E, Bolognesi, M. | | 登録日 | 2007-01-16 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Ferredoxin-NADP(+) Reductase from Plasmodium falciparum Undergoes NADP(+)-dependent Dimerization and Inactivation: Functional and Crystallographic Analysis.
J.Mol.Biol., 367, 2007
|
|
6CFJ
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with histidyl-CAM and bound to mRNA and A-, P-, and E-site tRNAs at 2.8A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | 登録日 | 2018-02-15 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
7RF9
 
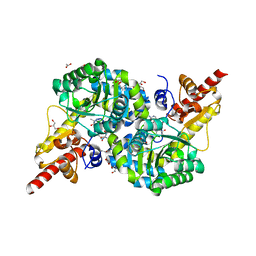 | | O2-, PLP-dependent desaturase Plu4 intermediate-bound enzyme | | 分子名称: | (2E)-5-carbamimidamido-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}pentanoic acid, 1,2-ETHANEDIOL, 2-(2-ETHOXYETHOXY)ETHANOL, ... | | 著者 | Hoffarth, E.R, Ryan, K.S. | | 登録日 | 2021-07-13 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.926 Å) | | 主引用文献 | A shared mechanistic pathway for pyridoxal phosphate-dependent arginine oxidases.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RGB
 
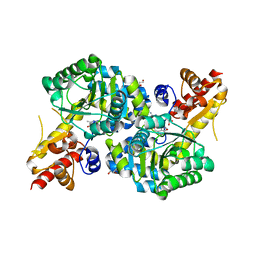 | | O2-, PLP-dependent desaturase Plu4 product-bound enzyme | | 分子名称: | (2Z,4E)-5-carbamimidamido-2-iminopent-4-enoic acid, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Hoffarth, E.R, Ryan, K.S. | | 登録日 | 2021-07-14 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A shared mechanistic pathway for pyridoxal phosphate-dependent arginine oxidases.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5NGK
 
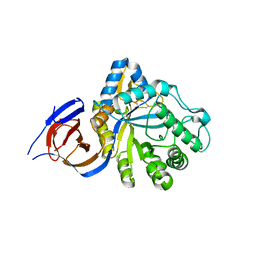 | | The endo-beta1,6-glucanase BT3312 | | 分子名称: | Glucosylceramidase | | 著者 | Basle, A, Temple, M, Cuskin, F, Lowe, E, Gilbert, H. | | 登録日 | 2017-03-17 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Bacteroidetes locus dedicated to fungal 1,6-beta-glucan degradation: Unique substrate conformation drives specificity of the key endo-1,6-beta-glucanase.
J. Biol. Chem., 292, 2017
|
|
1RFA
 
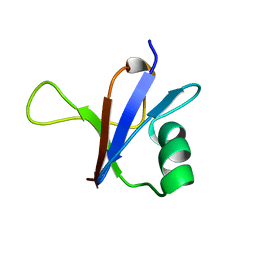 | | NMR SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF C-RAF-1 | | 分子名称: | RAF1 | | 著者 | Emerson, S.D, Madison, V.S, Palermo, R.E, Waugh, D.S, Scheffler, J.E, Tsao, K.-L, Kiefer, S.E, Liu, S.P, Fry, D.C. | | 登録日 | 1995-04-26 | | 公開日 | 1996-06-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Ras-binding domain of c-Raf-1 and identification of its Ras interaction surface.
Biochemistry, 34, 1995
|
|
5NIF
 
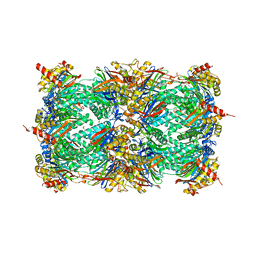 | | Yeast 20S proteasome in complex with Blm-pep activator | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Witkowska, J, Grudnik, P, Golik, P, Dubin, G, Jankowska, E. | | 登録日 | 2017-03-23 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of a low molecular weight activator Blm-pep with yeast 20S proteasome - insights into the enzyme activation mechanism.
Sci Rep, 7, 2017
|
|
7ZM7
 
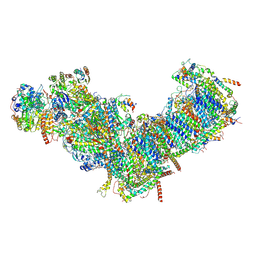 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Laube, E, Kuehlbrandt, W. | | 登録日 | 2022-04-19 | | 公開日 | 2022-11-30 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
3M2N
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{2-[N-(6-chloro-5-nitropyrimidin-4-yl)amino]ethyl}benzenesulfonamide | | 分子名称: | 4-{2-[(6-chloro-5-nitropyrimidin-4-yl)amino]ethyl}benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | 著者 | Grazulis, S, Manakova, E, Golovenko, D. | | 登録日 | 2010-03-08 | | 公開日 | 2011-02-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Design of [(2-pyrimidinylthio)acetyl]benzenesulfonamides as inhibitors of human carbonic anhydrases.
Eur.J.Med.Chem., 51, 2012
|
|
2XO5
 
 | | RIBONUCLEOTIDE REDUCTASE Y731NH2Y MODIFIED R1 SUBUNIT OF E. COLI | | 分子名称: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | 著者 | Minnihan, E.C, Seyedsayamdost, M.R, Uhlin, U, Stubbe, J. | | 登録日 | 2010-08-09 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Kinetics of Radical Intermediate Formation and Deoxynucleotide Production in 3-Aminotyrosine- Substituted Escherichia Coli Ribonucleotide Reductases.
J.Am.Chem.Soc., 133, 2011
|
|
6QCG
 
 | |
8FAI
 
 | | Cryo-EM structure of the Agrobacterium T-pilus | | 分子名称: | (7Z,19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacos-7-en-19-yl (9Z)-octadec-9-enoate, Protein virB2 | | 著者 | Kreida, S, Narita, A, Johnson, M.D, Tocheva, E.I, Das, A, Jensen, G.J, Ghosal, D. | | 登録日 | 2022-11-27 | | 公開日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structure of the Agrobacterium tumefaciens T4SS-associated T-pilus reveals stoichiometric protein-phospholipid assembly.
Structure, 31, 2023
|
|
7L7I
 
 | | Cryo-EM structure of Hsp90:FKBP51:p23 closed-state complex | | 分子名称: | Heat shock protein HSP 90-alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | 著者 | Lee, K, Thwin, A.C, Tse, E, Gates, S.N, Southworth, D.R. | | 登録日 | 2020-12-28 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state.
Mol.Cell, 81, 2021
|
|
7ZME
 
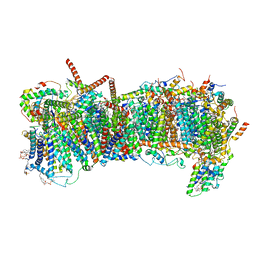 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) - membrane arm | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Laube, E, Kuehlbrandt, W. | | 登録日 | 2022-04-19 | | 公開日 | 2022-11-30 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
1PK7
 
 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with adenosine and sulfate/phosphate | | 分子名称: | ADENOSINE, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | 著者 | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | 登録日 | 2003-06-05 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
