6N57
 
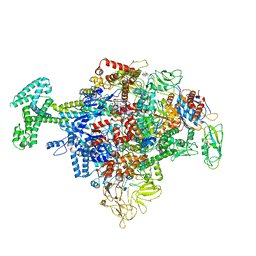 | | Cryo-EM structure of Escherichia coli RNAP polymerase bound with TraR in conformation I | | 分子名称: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | 登録日 | 2018-11-21 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
8CKB
 
 | | Asymmetric reconstruction of the crAss001 virion | | 分子名称: | Auxiliary capsid protein gp36, Cargo protein 1 gp45, Head fiber dimer protein gp29, ... | | 著者 | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | 登録日 | 2023-02-16 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.39 Å) | | 主引用文献 | Structural atlas of the most abundant human gut virus
Nature
|
|
1B3P
 
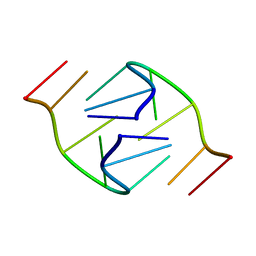 | | 5'-D(*GP*GP*AP*GP*GP*AP*T)-3' | | 分子名称: | DNA (5'-D(*GP*GP*AP*GP*GP*AP*T)-3') | | 著者 | Kettani, A, Bouaziz, S, Skripkin, E, Majumdar, A, Wang, W, Jones, R.A, Patel, D.J. | | 登録日 | 1998-12-14 | | 公開日 | 1999-08-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Interlocked mismatch-aligned arrowhead DNA motifs.
Structure Fold.Des., 7, 1999
|
|
1B6U
 
 | |
1B8K
 
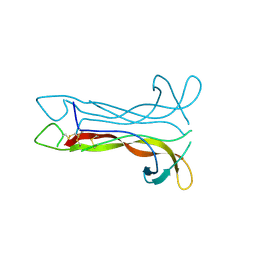 | | Neurotrophin-3 from Human | | 分子名称: | PROTEIN (NEUROTROPHIN-3) | | 著者 | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | 登録日 | 1999-02-01 | | 公開日 | 1999-02-09 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
6OYU
 
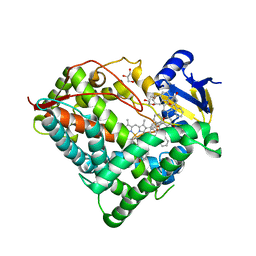 | | Structure of an ancestral-reconstructed cytochrome P450 1B1 with alpha-naphthoflavone | | 分子名称: | 2-PHENYL-4H-BENZO[H]CHROMEN-4-ONE, Cytochrome P450 1B1, GLYCEROL, ... | | 著者 | Bart, A.G, Harris, K.L, Scott, E.E. | | 登録日 | 2019-05-15 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structure of an ancestral mammalian family 1B1 cytochrome P450 with increased thermostability.
J.Biol.Chem., 295, 2020
|
|
8TKN
 
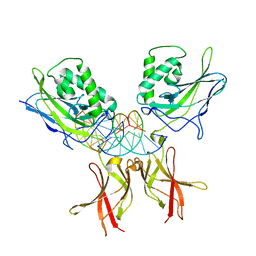 | | Murine NF-kappaB p50 Rel Homology Region homodimer in complex with 10-mer kappaB DNA from human Neutrophil Gelatinase-associated Lipocalin (NGAL) promoter | | 分子名称: | DNA A, DNA B, Nuclear factor NF-kappa-B p50 subunit | | 著者 | Zhu, N, Mealka, M, Mitchel, S, Rogers, W.E, Huxford, T. | | 登録日 | 2023-07-25 | | 公開日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Biomolecules, 13, 2023
|
|
6P07
 
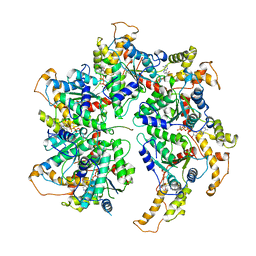 | | Spastin hexamer in complex with substrate | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Sandate, C.R, Szyk, A, Zehr, E, Roll-Mecak, A, Lander, G.C. | | 登録日 | 2019-05-16 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | An allosteric network in spastin couples multiple activities required for microtubule severing.
Nat.Struct.Mol.Biol., 26, 2019
|
|
1ATZ
 
 | |
1B2T
 
 | |
4UCS
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | 分子名称: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5-amino-3-(furan-2-yl)-1H-1,2,4-triazole-1-carboxamide, DNA LIGASE | | 著者 | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | 登録日 | 2014-12-04 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCO
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | 分子名称: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 7-amino-2-tert-butyl-4-(1H-pyrrol-2-yl)pyrido[2,3-d]pyrimidine-6-carboxamide, DNA LIGASE | | 著者 | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | 登録日 | 2014-12-04 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
6P9E
 
 | | Crystal structure of IL-36gamma complexed to A-552 | | 分子名称: | (2S)-2-{[4-(3-amino-4-methylphenyl)-6-methylpyrimidin-2-yl]oxy}-3-methoxy-3,3-diphenylpropanoic acid, Interleukin-36 gamma | | 著者 | Argiriadi, M.A, Todorovic, T, Su, Z, Putman, B, Kakavas, S.J, Salte, K.M, McDonald, H.A, Wetter, J.B, Paulsboe, S.E, Sun, Q, Gerstein, C.E, Medina, L, Sielaff, B, Sadhukhan, R, Stockmann, H, Richardson, P.L, Qiu, W, Henry, R.F, Herold, J.M, Shotwell, J.B, McGaraughty, S.P, Honore, P, Gopalakrishnan, S.M, Sun, C.C, Scott, V.E. | | 登録日 | 2019-06-10 | | 公開日 | 2019-06-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Small Molecule IL-36 gamma Antagonist as a Novel Therapeutic Approach for Plaque Psoriasis.
Sci Rep, 9, 2019
|
|
1BEG
 
 | | STRUCTURE OF FUNGAL ELICITOR, NMR, 18 STRUCTURES | | 分子名称: | BETA-ELICITIN CRYPTOGEIN | | 著者 | Fefeu, S, Bouaziz, S, Huet, J.-C, Pernollet, J.-C, Guittet, E. | | 登録日 | 1996-11-26 | | 公開日 | 1997-12-03 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of beta cryptogein, a beta elicitin secreted by a phytopathogenic fungus Phytophthora cryptogea.
Protein Sci., 6, 1997
|
|
8H86
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc | | 分子名称: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | 著者 | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | 登録日 | 2022-10-21 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.56 Å) | | 主引用文献 | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8H87
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc | | 分子名称: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR2, PALMITIC ACID, ... | | 著者 | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | 登録日 | 2022-10-21 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.53 Å) | | 主引用文献 | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8HTB
 
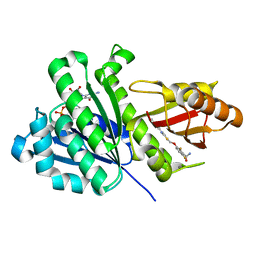 | | Staphylococcus aureus FtsZ 12-316 complexed with TXH9179 | | 分子名称: | 3-[(6-ethynyl-[1,3]thiazolo[5,4-b]pyridin-2-yl)methoxy]-2,6-bis(fluoranyl)benzamide, CALCIUM ION, Cell division protein FtsZ, ... | | 著者 | Bryan, E, Ferrer-Gonzalez, E, Sagong, H.Y, Fujita, J, Mark, L, Kaul, M, LaVoie, E.J, Matsumura, H, Pilch, D.S. | | 登録日 | 2022-12-21 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural and Antibacterial Characterization of a New Benzamide FtsZ Inhibitor with Superior Bactericidal Activity and In Vivo Efficacy Against Multidrug-Resistant Staphylococcus aureus.
Acs Chem.Biol., 18, 2023
|
|
4UTQ
 
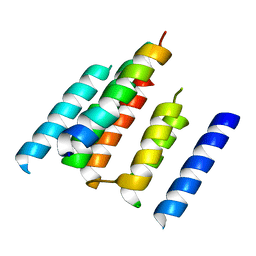 | | A structural model of the active ribosome-bound membrane protein insertase YidC | | 分子名称: | ATP SYNTHASE SUBUNIT C, MEMBRANE PROTEIN INSERTASE YIDC | | 著者 | Wickles, S, Singharoy, A, Andreani, J, Seemayer, S, Bischoff, L, Berninghausen, O, Soeding, J, Schulten, K, vanderSluis, E.O, Beckmann, R. | | 登録日 | 2014-07-22 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | A Structural Model of the Active Ribosome-Bound Membrane Protein Insertase Yidc.
Elife, 3, 2014
|
|
6X9F
 
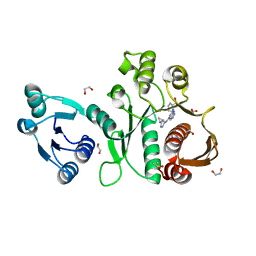 | | Pseudomonas aeruginosa MurC with AZ8074 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Horanyi, P.S, Mayclin, S.J, Durand-Reville, T.F, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2020-06-02 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Pseudomonas aeruginosa MurC with AZ8074
To Be Published
|
|
6N2W
 
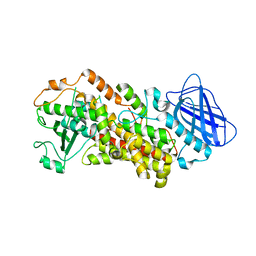 | | The structure of Stable-5-Lipoxygenase bound to NDGA | | 分子名称: | 4-[(2R,3S)-3-[(3,4-DIHYDROXYPHENYL)METHYL]-2-METHYLBUTYL]BENZENE-1,2-DIOL, Arachidonate 5-lipoxygenase, FE (II) ION | | 著者 | Newcomer, M.E, Gilbert, N.C, Neau, D.B. | | 登録日 | 2018-11-14 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structural and mechanistic insights into 5-lipoxygenase inhibition by natural products.
Nat.Chem.Biol., 16, 2020
|
|
1B1V
 
 | | NMR STRUCTURE OF PSP1, PLASMATOCYTE-SPREADING PEPTIDE FROM PSEUDOPLUSIA INCLUDENS | | 分子名称: | PROTEIN (PLASMATOCYTE-SPREADING PEPTIDE) | | 著者 | Volkman, B.F, Clark, K.D, Anderson, M.E, Pech, L.L, Markley, J.L, Strand, M.R. | | 登録日 | 1998-11-23 | | 公開日 | 1998-12-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the insect cytokine peptide plasmatocyte-spreading peptide 1 from Pseudoplusia includens.
J.Biol.Chem., 274, 1999
|
|
7N6I
 
 | | ATP-bound TnsC-TniQ complex from ShCAST system | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | 著者 | Park, J, Tsai, A.W.L, Mehrotra, E, Kellogg, E.H. | | 登録日 | 2021-06-08 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for target site selection in RNA-guided DNA transposition systems.
Science, 373, 2021
|
|
7OJK
 
 | | Lassa virus L protein bound to the distal promoter duplex [DISTAL-PROMOTER] | | 分子名称: | 3' RNA, 5' RNA, RNA-directed RNA polymerase L, ... | | 著者 | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | 登録日 | 2021-05-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.89 Å) | | 主引用文献 | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
7OJL
 
 | | Lassa virus L protein in a pre-initiation conformation [PREINITIATION] | | 分子名称: | 3' RNA, 5' RNA, MANGANESE (II) ION, ... | | 著者 | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | 登録日 | 2021-05-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
1AXU
 
 | | SOLUTION NMR STRUCTURE OF THE [AP]DG ADDUCT OPPOSITE DA IN A DNA DUPLEX, NMR, 9 STRUCTURES | | 分子名称: | DNA DUPLEX D(CCATC-[AP]G-CTACC)D(GGTAGAGATGG), N-1-AMINOPYRENE | | 著者 | Gu, Z, Gorin, A.A, Krishnasami, R, Hingerty, B.E, Basu, A.K, Broyde, S, Patel, D.J. | | 登録日 | 1997-10-21 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dA in a DNA duplex.
Biochemistry, 38, 1999
|
|
