8KD0
 
 | |
6R2N
 
 | | Crystal structure of KlGlk1 glucokinase from Kluyveromyces lactis | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Glucokinase-1 | | Authors: | Zak, K, Wator, E, Grudnik, P. | | Deposit date: | 2019-03-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Crystal Structure of Kluyveromyces lactis Glucokinase ( Kl Glk1).
Int J Mol Sci, 20, 2019
|
|
6BNA
 
 | | BINDING OF AN ANTITUMOR DRUG TO DNA. NETROPSIN AND C-G-C-G-A-A-T-T-BRC-G-C-G | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(CBR)P*GP*CP*G)-3'), NETROPSIN | | Authors: | Kopka, M.L, Yoon, C, Goodsell, D, Pjura, P, Dickerson, R.E. | | Deposit date: | 1984-08-30 | | Release date: | 1984-10-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G.
J.Mol.Biol., 183, 1985
|
|
5YTN
 
 | | C135A mutant of copper-containing nitrite reductase from Geobacillus thermodenitrificans in complex with peroxide | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Fukuda, Y, Matsusaki, T, Tse, K.M, Mizohata, E, Murphy, M.E.P, Inoue, T. | | Deposit date: | 2017-11-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic study of dioxygen chemistry in a copper-containing nitrite reductase from Geobacillus thermodenitrificans.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6C23
 
 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-05 | | Release date: | 2018-01-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
6FD7
 
 | |
6F8R
 
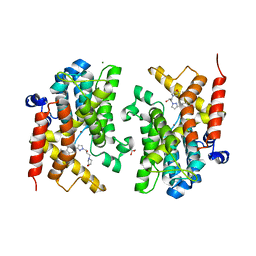 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-54 | | Descriptor: | (2~{S})-1-[3-(3-cyclopentyloxy-4-methoxy-phenyl)pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.826 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6H5D
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 2 | | Descriptor: | (1~{S},3~{S},4~{R},5~{R})-3-methyl-1,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 9, 2019
|
|
5J1G
 
 | |
4PA9
 
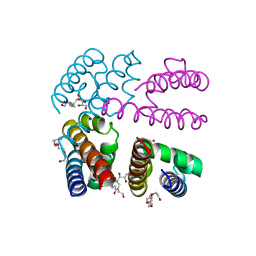 | | Structure of NavMS in complex with channel blocking compound | | Descriptor: | BROMIDE ION, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3KDB
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10006 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Chufan, E.E, Lafont, V, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
1V0H
 
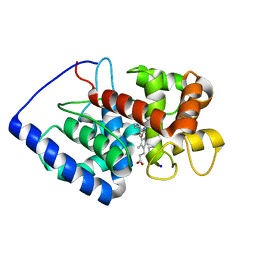 | | ASCOBATE PEROXIDASE FROM SOYBEAN CYTOSOL IN COMPLEX WITH SALICYLHYDROXAMIC ACID | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SALICYLHYDROXAMIC ACID, ... | | Authors: | Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2004-03-29 | | Release date: | 2004-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Ascorbate Peroxidase-Salicylhydroxamic Acid Complex
Biochemistry, 43, 2004
|
|
1UXU
 
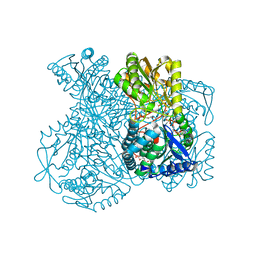 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
6FDC
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-32a | | Descriptor: | (2~{R})-1-[3-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, (2~{S})-1-[5-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, 1,2-ETHANEDIOL, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
4PAV
 
 | | Structure of hypothetical protein SA1046 from S. aureus. | | Descriptor: | Glyoxalase family protein | | Authors: | Battaile, K.P, Mulichak, A, Lam, R, Lam, K, Soloveychik, M, Romanov, V, Jones, K, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of hypothetical protein SA1046 from S. aureus.
To Be Published
|
|
6GGR
 
 | | Crystal structure of Salmonella zinc metalloprotease effector GtgA in complex with p65 | | Descriptor: | Bacteriophage virulence determinant, CHLORIDE ION, Transcription factor p65 | | Authors: | Jennings, E, Esposito, D, Rittinger, K, Thurston, T. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure-function analyses of the bacterial zinc metalloprotease effector protein GtgA uncover key residues required for deactivating NF-kappa B.
J. Biol. Chem., 293, 2018
|
|
2HO4
 
 | | Crystal Structure of Protein from Mouse Mm.236127 | | Descriptor: | Haloacid dehalogenase-like hydrolase domain containing 2, MAGNESIUM ION, PHOSPHATE ION | | Authors: | McCoy, J.G, Wesenberg, G.E, Bitto, E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-07-13 | | Release date: | 2006-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Protein from Mouse Mm.236127
To be published
|
|
7UTY
 
 | | First bromodomain of BRD4 liganded with compound 2c | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, prop-2-en-1-yl (5S)-1-ethyl-7-methyl-5-(4-methylphenyl)-2,4-dioxo-1,2,3,4,5,8-hexahydropyrido[2,3-d]pyrimidine-6-carboxylate | | Authors: | Schonbrunn, E, Chan, A. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1,4-Dihydropyridinebutyrolactone-derived ring-opened ester and amide analogs targeting BET bromodomains.
Arch Pharm, 355, 2022
|
|
5IHH
 
 | | Crystal structure of human transthyretin in complex with luteolin-MeO at 1.35 A resolution | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5-hydroxy-7-methoxy-4H-1-chromen-4-one, SODIUM ION, Transthyretin | | Authors: | Begum, A, Nilsson, L, Olofsson, A, Sauer-Eriksson, A.E. | | Deposit date: | 2016-02-29 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Modifications of the 7-Hydroxyl Group of the Transthyretin Ligand Luteolin Provide Mechanistic Insights into Its Binding Properties and High Plasma Specificity.
Plos One, 11, 2016
|
|
6C73
 
 | | Tryptophan synthase Q114A mutant (internal aldimine state) in complex with N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) with cesium ion bound in the metal coordination site | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, CESIUM ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2018-01-19 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Tryptophan synthase Q114A mutant (internal aldimine state) in complex with N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) with cesium ion bound in the metal coordination site.
To be Published
|
|
4PEI
 
 | | Dbr1 in complex with synthetic branched RNA analog | | Descriptor: | GLYCEROL, NICKEL (II) ION, RNA (5'-R(*(G46)P*U)-3'), ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
4PEY
 
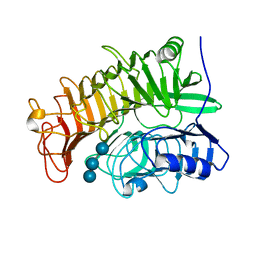 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose | | Descriptor: | Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
1V8P
 
 | | Crystal structure of PAE2754 from Pyrobaculum aerophilum | | Descriptor: | CHLORIDE ION, hypothetical protein PAE2754 | | Authors: | Arcus, V.L, Backbro, K, Roos, A, Daniel, E.L, Baker, E.N. | | Deposit date: | 2004-01-12 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease
J.Biol.Chem., 279, 2004
|
|
6TTJ
 
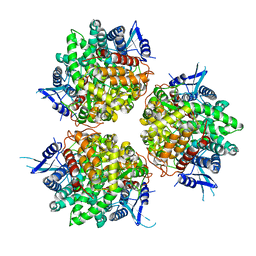 | | Neutral invertase 2 from Arabidopsis thaliana | | Descriptor: | Alkaline/neutral invertase CINV1 | | Authors: | Tsirkone, V.G, Osipov, E.M, Beelen, S, Strelkov, S.V. | | Deposit date: | 2019-12-27 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.392 Å) | | Cite: | Crystal structure of Arabidopsis thaliana neutral invertase 2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6MMW
 
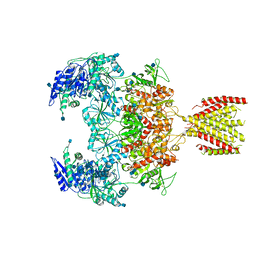 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2A* in the '2-Knuckle-Symmetric' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-10-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
