3LA5
 
 | |
6Q57
 
 | |
3GYQ
 
 | |
3JSX
 
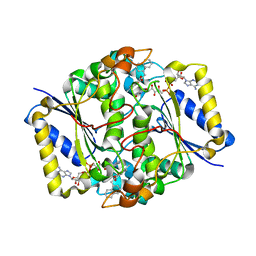 | | X-ray Crystal structure of NAD(P)H: Quinone Oxidoreductase-1 (NQO1) bound to the coumarin-based inhibitor AS1 | | 分子名称: | 4-hydroxy-6,7-dimethyl-3-(naphthalen-1-ylmethyl)-2H-chromen-2-one, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | 著者 | Dunstan, M.S, Levy, C, Leys, D. | | 登録日 | 2009-09-11 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Synthesis and biological evaluation of coumarin-based inhibitors of NAD(P)H: quinone oxidoreductase-1 (NQO1).
J.Med.Chem., 52, 2009
|
|
1Y39
 
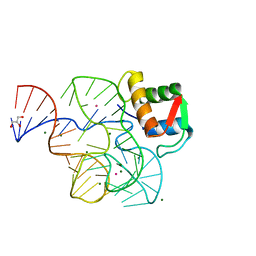 | | Co-evolution of protein and RNA structures within a highly conserved ribosomal domain | | 分子名称: | 50S ribosomal protein L11, 58 Nucleotide Ribosomal 23S RNA Domain, COBALT (III) ION, ... | | 著者 | Dunstan, M.S, GuhaThakurta, D, Draper, D.E, Conn, G.L. | | 登録日 | 2004-11-24 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Coevolution of Protein and RNA Structures within a Highly Conserved Ribosomal Domain
Chem.Biol., 12, 2005
|
|
6GKZ
 
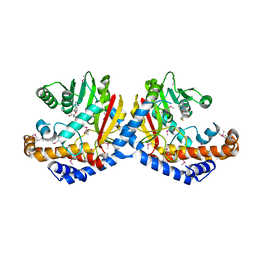 | |
6GKY
 
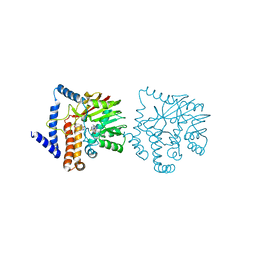 | |
4EPP
 
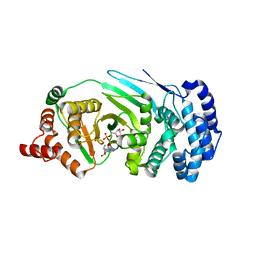 | |
4EPQ
 
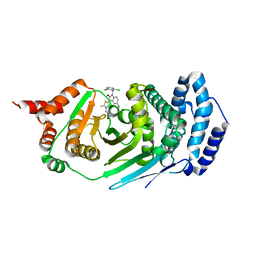 | |
4LX5
 
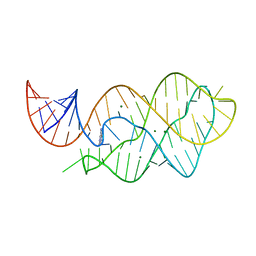 | | X-ray crystal structure of the M6" riboswitch aptamer bound to pyrimido[4,5-d]pyrimidine-2,4-diamine (PPDA) | | 分子名称: | MAGNESIUM ION, Mutated adenine riboswitch aptamer, pyrimido[4,5-d]pyrimidine-2,4-diamine | | 著者 | Dunstan, M.S, Leys, D. | | 登録日 | 2013-07-29 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Modular riboswitch toolsets for synthetic genetic control in diverse bacterial species.
J.Am.Chem.Soc., 136, 2014
|
|
4LX6
 
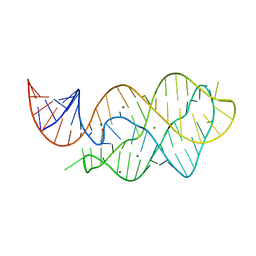 | |
6GKV
 
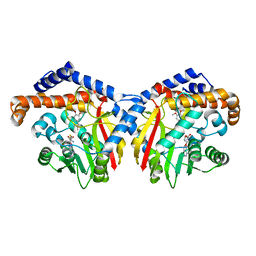 | | Crystal structure of Coclaurine N-Methyltransferase (CNMT) bound to N-methylheliamine and SAH | | 分子名称: | 6,7-dimethoxy-2-methyl-1,2,3,4-tetrahydroisoquinolin-2-ium, Coclaurine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Dunstan, M.S, Levy, C.W. | | 登録日 | 2018-05-22 | | 公開日 | 2018-06-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure and Biocatalytic Scope of Coclaurine N-Methyltransferase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3SII
 
 | |
3SIJ
 
 | |
3SIH
 
 | |
3TE7
 
 | | Quinone Oxidoreductase (NQ02) bound to the imidazoacridin-6-one 5a1 | | 分子名称: | 5-{[2-(dimethylamino)ethyl]amino}-8-methoxy-6H-imidazo[4,5,1-de]acridin-6-one, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | 著者 | Dunstan, M.S. | | 登録日 | 2011-08-12 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Novel Inhibitors of NRH:Quinone Oxidoreductase 2 (NQO2): Crystal Structures, Biochemical Activity, and Intracellular Effects of Imidazoacridin-6-ones.
J.Med.Chem., 54, 2011
|
|
3TEM
 
 | | Quinone Oxidoreductase (NQ02) bound to the imidazoacridin-6-one 6a1 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | 著者 | Dunstan, M.S, Leys, D. | | 登録日 | 2011-08-15 | | 公開日 | 2011-09-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Novel Inhibitors of NRH:Quinone Oxidoreductase 2 (NQO2): Crystal Structures, Biochemical Activity, and Intracellular Effects of Imidazoacridin-6-ones.
J.Med.Chem., 54, 2011
|
|
3TZB
 
 | | Quinone Oxidoreductase (NQ02) bound to NSC13000 | | 分子名称: | 9-AMINOACRIDINE, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | 著者 | Dunstan, M.S, Leys, D. | | 登録日 | 2011-09-27 | | 公開日 | 2011-11-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1901 Å) | | 主引用文献 | In silico screening reveals structurally diverse, nanomolar inhibitors of NQO2 that are functionally active in cells and can modulate NF-kappa B signaling.
Mol.Cancer Ther., 11, 2012
|
|
5LV9
 
 | |
5LOA
 
 | |
5LTM
 
 | |
5LOC
 
 | |
5LVA
 
 | |
5MSD
 
 | |
5MSC
 
 | |
