4V9D
 
 | | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Wang, L, Feldman, M.B, Pulk, A, Chen, V.B, Kapral, G.J, Noeske, J, Richardson, J.S, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2012-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding.
Science, 332, 2011
|
|
9D2D
 
 | |
8VBS
 
 | |
6UY5
 
 | |
4V7T
 
 | | Crystal structure of the E. coli ribosome bound to chloramphenicol. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-14 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1942 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V7S
 
 | | Crystal structure of the E. coli ribosome bound to telithromycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-05 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2547 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V7U
 
 | | Crystal structure of the E. coli ribosome bound to erythromycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4WWW
 
 | | Crystal structure of the E. coli ribosome bound to CEM-101 | | Descriptor: | (3aS,4R,7S,9R,10R,11R,13R,15R,15aR)-1-{4-[4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl]butyl}-4-ethyl-7-fluoro-11-methoxy-3a ,7,9,11,13,15-hexamethyl-2,6,8,14-tetraoxotetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-10-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S rRNA, 23S rRNA, ... | | Authors: | Dunkle, J.A, Zhang, W, Cate, J.H.D, Mankin, A.S. | | Deposit date: | 2014-11-12 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Binding and action of CEM-101, a new fluoroketolide antibiotic that inhibits protein synthesis.
Antimicrob. Agents Chemother., 54, 2010
|
|
7RRN
 
 | |
7RUJ
 
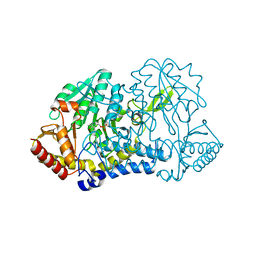 | | E. coli cysteine desulfurase SufS N99A | | Descriptor: | CHLORIDE ION, Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dunkle, J.A, Gogar, R, Frantom, P.A. | | Deposit date: | 2021-08-17 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The beta-latch structural element of the SufS cysteine desulfurase mediates active site accessibility and SufE transpersulfurase positioning.
J.Biol.Chem., 299, 2023
|
|
7RW3
 
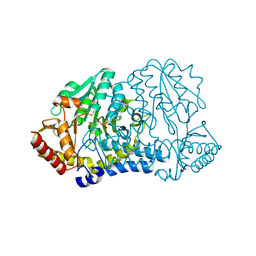 | | E. coli cysteine desulfurase SufS N99D | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dunkle, J.A, Gogar, R, Frantom, P.A. | | Deposit date: | 2021-08-19 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The beta-latch structural element of the SufS cysteine desulfurase mediates active site accessibility and SufE transpersulfurase positioning.
J.Biol.Chem., 299, 2023
|
|
4OX9
 
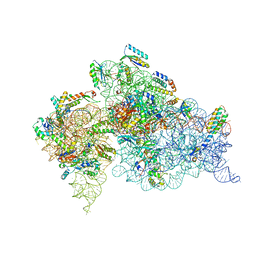 | | Crystal structure of the aminoglycoside resistance methyltransferase NpmA bound to the 30S ribosomal subunit | | Descriptor: | 16S rRNA, 16S rRNA (adenine(1408)-N(1))-methyltransferase, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Conn, G.L, Dunham, C.M. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8035 Å) | | Cite: | Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistance methyltransferase NpmA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4V7V
 
 | | Crystal structure of the E. coli ribosome bound to clindamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-16 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.2891 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6MRH
 
 | |
6MRE
 
 | |
6MRI
 
 | |
6MR2
 
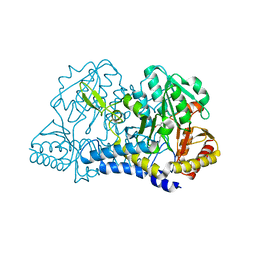 | |
6MR6
 
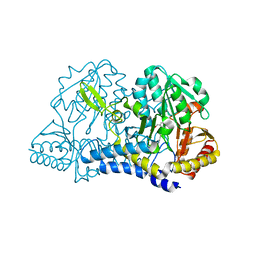 | |
6O12
 
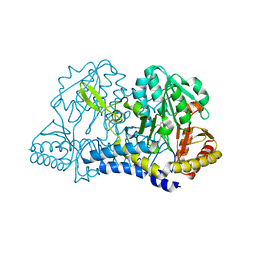 | | E. coli cysteine desulfurase SufS H123A | | Descriptor: | CHLORIDE ION, Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dunkle, J.A, Frantom, P.A. | | Deposit date: | 2019-02-17 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Direct observation of intermediates in the SufS cysteine desulfurase reaction reveals functional roles of conserved active-site residues.
J.Biol.Chem., 294, 2019
|
|
6O13
 
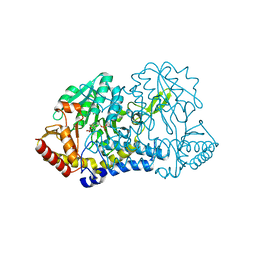 | |
6O10
 
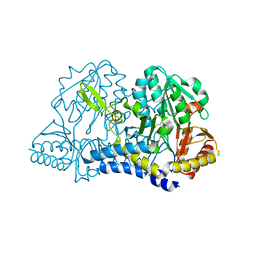 | | E. coli cysteine desulfurase SufS | | Descriptor: | CHLORIDE ION, Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dunkle, J.A, Frantom, P.A. | | Deposit date: | 2019-02-17 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct observation of intermediates in the SufS cysteine desulfurase reaction reveals functional roles of conserved active-site residues.
J.Biol.Chem., 294, 2019
|
|
6O11
 
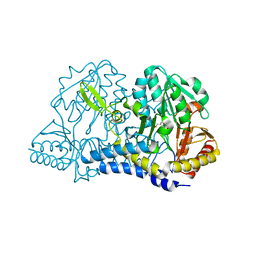 | | E. coli cysteine desulfurase SufS C364A with a Cys-aldimine intermediate | | Descriptor: | CHLORIDE ION, Cysteine desulfurase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Dunkle, J.A, Frantom, P.A. | | Deposit date: | 2019-02-17 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Direct observation of intermediates in the SufS cysteine desulfurase reaction reveals functional roles of conserved active-site residues.
J.Biol.Chem., 294, 2019
|
|
8DO6
 
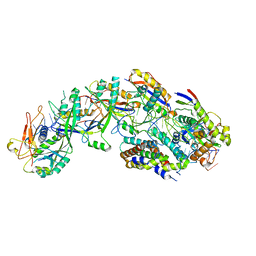 | | The structure of S. epidermidis Cas10-Csm bound to target RNA | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Paraan, M, Stagg, S.M, Dunkle, J.A. | | Deposit date: | 2022-07-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of a Type III-A CRISPR-Cas effector complex reveals conserved and idiosyncratic contacts to target RNA and crRNA among Type III-A systems.
Plos One, 18, 2023
|
|
1VVJ
 
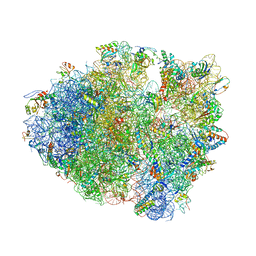 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 bound to Codon CCC-G on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-05-24 | | Release date: | 2014-08-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.440001 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4V6E
 
 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.712 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
