7WRJ
 
 | |
7V20
 
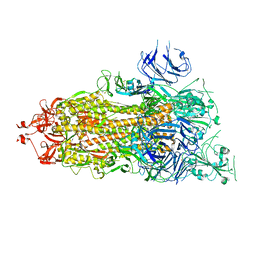 | | CryoEM structure of del68-76/del679-688 prefusion-stabilized spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Du, S, Xiao, J. | | Deposit date: | 2021-08-07 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | CryoEM structure of del68-76/del679-688 prefusion-stabilized spike
To Be Published
|
|
7V23
 
 | |
7UY0
 
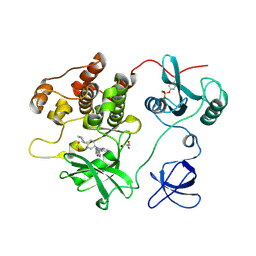 | | Crystal structure of human Fgr tyrosine kinase in complex with A-419259 | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CHLORIDE ION, GLYCEROL, ... | | Authors: | Du, S, Alvarado, J.J, Smithgall, T.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | ATP-site inhibitors induce unique conformations of the acute myeloid leukemia-associated Src-family kinase, Fgr.
Structure, 30, 2022
|
|
7V22
 
 | |
7V24
 
 | |
7WR9
 
 | |
7WCK
 
 | |
7WCU
 
 | |
7WCH
 
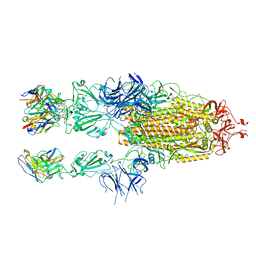 | |
7WCP
 
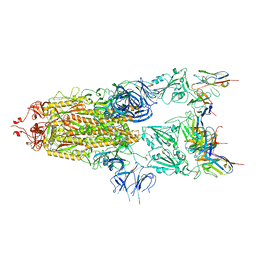 | |
7UY3
 
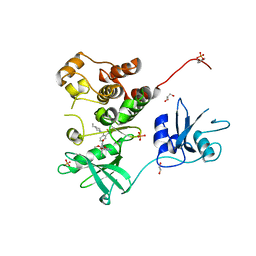 | | Crystal structure of human Fgr tyrosine kinase in complex with TL02-59 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(6,7-dimethoxyquinazolin-4-yl)oxy]-N-{4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-4-methylbenzamide, GLYCEROL, ... | | Authors: | Du, S, Alvarado, J.J, Smithgall, T.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | ATP-site inhibitors induce unique conformations of the acute myeloid leukemia-associated Src-family kinase, Fgr.
Structure, 30, 2022
|
|
1M5W
 
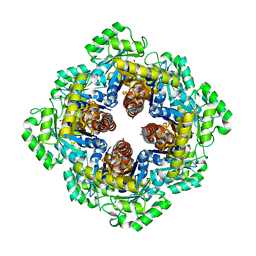 | | 1.96 A Crystal Structure of Pyridoxine 5'-Phosphate Synthase in Complex with 1-deoxy-D-xylulose phosphate | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, PHOSPHATE ION, Pyridoxal phosphate biosynthetic protein pdxJ | | Authors: | Yeh, J.I, Du, S, Pohl, E, Cane, D.E. | | Deposit date: | 2002-07-10 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Multistate Binding in Pyridoxine 5'-Phosphate Synthase: 1.96 A Crystal Structure in
Complex with 1-deoxy-D-xylulose phosphate
Biochemistry, 41, 2002
|
|
2R4E
 
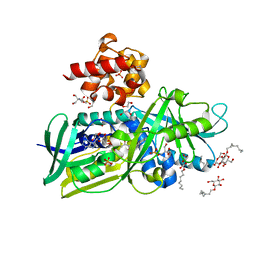 | | Crystal structure of Escherichia coli Glycerol-3-phosphate Dehydrogenase in complex with DHAP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, Aerobic glycerol-3-phosphate dehydrogenase, ... | | Authors: | Yeh, J.I, Du, S, Chinte, U. | | Deposit date: | 2007-08-31 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2R45
 
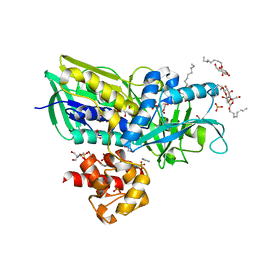 | | Crystal structure of Escherichia coli Glycerol-3-phosphate Dehydrogenase in complex with 2-phospho-d-glyceric acid | | Descriptor: | 1,2-ETHANEDIOL, 2-PHOSPHOGLYCERIC ACID, Aerobic glycerol-3-phosphate dehydrogenase, ... | | Authors: | Yeh, J.I, Du, S, Chinte, U. | | Deposit date: | 2007-08-30 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential
monotopic membrane enzyme involved in respiration and metabolism
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2R46
 
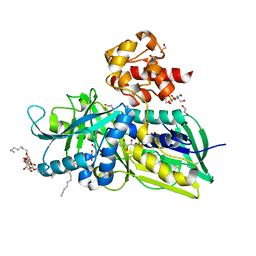 | | Crystal structure of Escherichia coli Glycerol-3-phosphate Dehydrogenase in complex with 2-phosphopyruvic acid. | | Descriptor: | 1,2-ETHANEDIOL, Aerobic glycerol-3-phosphate dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yeh, J.I, Du, S, Chinte, U. | | Deposit date: | 2007-08-30 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential
monotopic membrane enzyme involved in respiration and metabolism
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3PA0
 
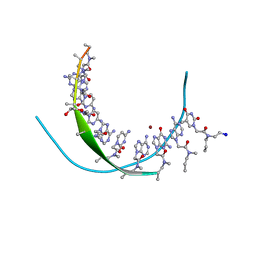 | | Crystal Structure of Chiral Gamma-PNA with Complementary DNA Strand: Insight Into the Stability and Specificity of Recognition an Conformational Preorganization | | Descriptor: | 1,2-ETHANEDIOL, DNA 5'-D(*AP*TP*CP*TP*GP*TP*GP*GP*TP*C)-3', PEPTIDE NUCLEIC ACID, ... | | Authors: | Yeh, J.I, Shivachev, B, Du, S. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of chiral gammaPNA with complementary DNA strand: insights into the stability and specificity of recognition and conformational preorganization.
J.Am.Chem.Soc., 132, 2010
|
|
5GJA
 
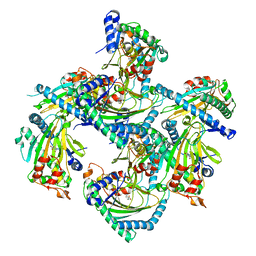 | | Crystal structure of Arabidopsis thaliana ACO2 in complex with 2-PA | | Descriptor: | 1-aminocyclopropane-1-carboxylate oxidase 2, PYRIDINE-2-CARBOXYLIC ACID, ZINC ION | | Authors: | Sun, X.Z, Li, Y.X, He, W.R, Ji, C.G, Xia, P.X, Wang, Y.C, Du, S, Li, H.J, Raikhel, N, Xiao, J.Y, Guo, H.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase.
Nat Commun, 8, 2017
|
|
5GJ9
 
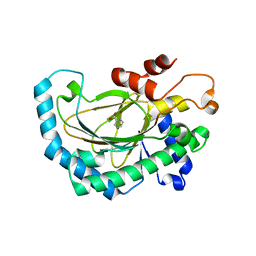 | | Crystal structure of Arabidopsis thaliana ACO2 in complex with POA | | Descriptor: | 1-aminocyclopropane-1-carboxylate oxidase 2, PYRAZINE-2-CARBOXYLIC ACID, ZINC ION | | Authors: | Sun, X.Z, Li, Y.X, He, W.R, Ji, C.G, Xia, P.X, Wang, Y.C, Du, S, Li, H.J, Raikhel, N, Xiao, J.Y, Guo, H.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase.
Nat Commun, 8, 2017
|
|
3MBU
 
 | | Structure of a bipyridine-modified PNA duplex | | Descriptor: | 1,2-ETHANEDIOL, Bipyridine-PNA, CARBONATE ION, ... | | Authors: | Yeh, J.I, Pohl, E, Truan, D, He, W, Sheldrick, G.M, Du, S, Achim, C. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystal structure of non-modified and bipyridine-modified PNA duplexes.
Chemistry, 16, 2010
|
|
4E5Z
 
 | |
4E54
 
 | |
2QCU
 
 | | Crystal structure of Glycerol-3-phosphate Dehydrogenase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Aerobic glycerol-3-phosphate dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yeh, J.I, Chinte, U, Du, S. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2R4J
 
 | | Crystal structure of Escherichia coli SeMet substituted Glycerol-3-phosphate Dehydrogenase in complex with DHAP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, Aerobic glycerol-3-phosphate dehydrogenase, ... | | Authors: | Yeh, J.I, Du, S, Chinte, U. | | Deposit date: | 2007-08-31 | | Release date: | 2008-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
