3LZQ
 
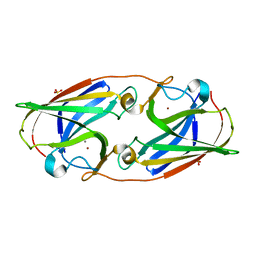 | | Crystal Structure Analysis of Manganese treated P19 protein from Campylobacter jejuni at 1.41 A at pH 9 | | Descriptor: | COPPER (II) ION, MANGANESE (II) ION, P19 protein, ... | | Authors: | Doukov, T.I, Chan, A.C.K, Scofield, M, Ramin, A.B, Tom-Yew, S.A.L, Murphy, M.E.P. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure and Function of P19, a High-Affinity Iron Transporter of the Human Pathogen Campylobacter jejuni.
J.Mol.Biol., 401, 2010
|
|
3LZR
 
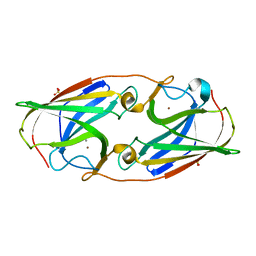 | | Crystal Structure Analysis of Manganese treated P19 protein from Campylobacter jejuni at 2.73 A at pH 9 and Manganese peak wavelength (1.893 A) | | Descriptor: | COPPER (II) ION, MANGANESE (II) ION, P19 protein, ... | | Authors: | Doukov, T.I, Chan, A.C.K, Scofield, M, Ramin, A.B, Tom-Yew, S.A.L, Murphy, M.E.P. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure and Function of P19, a High-Affinity Iron Transporter of the Human Pathogen Campylobacter jejuni.
J.Mol.Biol., 401, 2010
|
|
7UR2
 
 | |
4GED
 
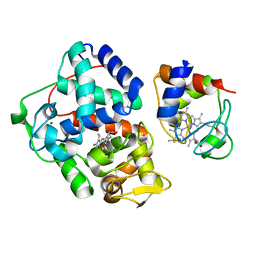 | | Crystal Structure of the Leishmania Major Peroxidase-Cytochrome C Complex | | Descriptor: | Ascorbate peroxidase, Cytochrome c, HEME C, ... | | Authors: | Jasion, V.S, Doukov, T, Pineda, S.H, Li, H, Poulos, T.L. | | Deposit date: | 2012-08-01 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the Leishmania major peroxidase-cytochrome c complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
9CPL
 
 | |
7N1O
 
 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Transcription factor ETV6,Isoform 4 of Anthrax toxin receptor 2 | | Authors: | Mathis, M.H, Bezzant, B.D, Ramirez, D.T, Sarath Nawarathnage, S.D, Doukov, T, Moody, J.D. | | Deposit date: | 2021-05-27 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystals of TELSAM-target protein fusions that exhibit minimal crystal contacts and lack direct inter-TELSAM contacts.
Open Biology, 12, 2022
|
|
7N2B
 
 | | A DARPin semi-rigidly fused to the 3TEL crystallization chaperone | | Descriptor: | Transcription factor ETV6,Transcription factor ETV6,Transcription factor ETV6,3TEL-rigid-DARPin | | Authors: | Sarath Nawarathange, S.D, Gajjar, P, Bunn, D, Stewart, C, Doukov, T, Moody, J.D. | | Deposit date: | 2021-05-28 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.221 Å) | | Cite: | Crystals of TELSAM-target protein fusions that exhibit minimal crystal contacts and lack direct inter-TELSAM contacts.
Open Biology, 12, 2022
|
|
8ASH
 
 | | Crystal structure of d(CCGGGGTACCCCGG) with XRB | | Descriptor: | 4-[(~{E})-(3,6-dimethyl-1,3-benzothiazol-2-yl)iminomethyl]-~{N},~{N}-dimethyl-aniline, DNA (5'-D(*CP*CP*GP*GP*GP*GP*TP*AP*CP*CP*CP*CP*GP*G)-3') | | Authors: | Sbirkova-Dimitrova, H.I, Shivachev, B.L, Rusev, R, Kuvandjiev, N, Heroux, A, Doukov, T. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.837 Å) | | Cite: | Structural Characterization of Alzheimer DNA Promoter Sequences from the Amyloid Precursor Gene in the Presence of Thioflavin T and Analogs
Crystals, 12, 2022
|
|
8ASK
 
 | | Crystal structure of d(GCCCACCACGGC) | | Descriptor: | DNA (5'-D(P*GP*CP*CP*CP*AP*CP*CP*AP*CP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*GP*TP*GP*GP*TP*GP*GP*GP*C)-3') | | Authors: | Sbirkova-Dimitrova, H.I, Shivachev, B.L, Rusev, R, Heroux, A, Doukov, T, Kuvandjiev, N. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Structural Characterization of Alzheimer DNA Promoter Sequences from the Amyloid Precursor Gene in the Presence of Thioflavin T and Analogs
Crystals, 12, 2022
|
|
6MKB
 
 | | Crystal structure of murine 4-1BB ligand | | Descriptor: | SODIUM ION, SULFATE ION, Tumor necrosis factor ligand superfamily member 9, ... | | Authors: | Bitra, A, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-09-25 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the m4-1BB/4-1BBL complex reveals an unusual dimeric ligand that undergoes structural changes upon 4-1BB receptor binding.
J. Biol. Chem., 294, 2019
|
|
6MKZ
 
 | | Crystal structure of murine 4-1BB/4-1BBL complex | | Descriptor: | Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bitra, A, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-09-26 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the m4-1BB/4-1BBL complex reveals an unusual dimeric ligand that undergoes structural changes upon 4-1BB receptor binding.
J. Biol. Chem., 294, 2019
|
|
5F4H
 
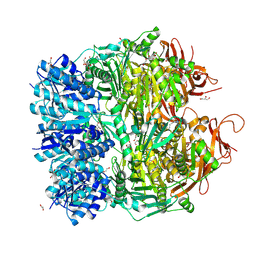 | | Archael RuvB-like Holiday junction helicase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleotide binding protein PINc | | Authors: | Zhai, B, DuPrez, K.T, Doukov, T.I, Shen, Y, Fan, L. | | Deposit date: | 2015-12-03 | | Release date: | 2016-12-21 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structure and Function of a Novel ATPase that Interacts with Holliday Junction Resolvase Hjc and Promotes Branch Migration.
J. Mol. Biol., 429, 2017
|
|
5WJF
 
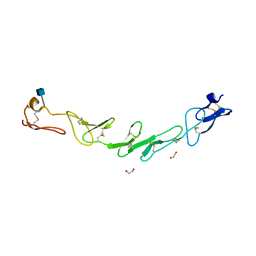 | | Crystal structure of murine 4-1BB from HEK293T cells in P21212 space group | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor receptor superfamily member 9 | | Authors: | Zajonc, D.M, Doukov, T, Bitra, A. | | Deposit date: | 2017-07-21 | | Release date: | 2017-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of murine 4-1BB and its interaction with 4-1BBL support a role for galectin-9 in 4-1BB signaling.
J. Biol. Chem., 293, 2018
|
|
6NYP
 
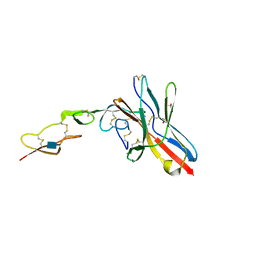 | | Crystal structure of UL144/BTLA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B- and T-lymphocyte attenuator, GLYCEROL, ... | | Authors: | Aruna, B, Zajonc, D.M, Doukov, T. | | Deposit date: | 2019-02-11 | | Release date: | 2019-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of human cytomegalovirus UL144, an HVEM orthologue, bound to the B and T cell lymphocyte attenuator.
J.Biol.Chem., 294, 2019
|
|
4DJD
 
 | | Crystal structure of folate-free corrinoid iron-sulfur protein (CFeSP) in complex with its methyltransferase (MeTr) | | Descriptor: | 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase, CALCIUM ION, COBALAMIN, ... | | Authors: | Kung, Y, Doukov, T.I, Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2012-02-01 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Visualizing molecular juggling within a B12-dependent methyltransferase complex.
Nature, 484, 2012
|
|
6OFB
 
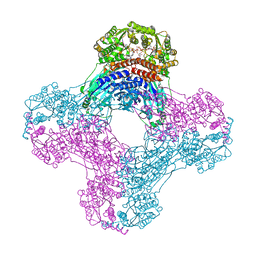 | | Crystal structure of human glutamine-dependent NAD+ synthetase complexed with NaAD+, AMP, pyrophosphate, and Mg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Glutamine-dependent NAD(+) synthetase, ... | | Authors: | Chuenchor, W, Doukov, T.I, Gerratana, B. | | Deposit date: | 2019-03-28 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Different ways to transport ammonia in human and Mycobacterium tuberculosis NAD+synthetases.
Nat Commun, 11, 2020
|
|
6OFC
 
 | | Crystal structure of M. tuberculosis glutamine-dependent NAD+ synthetase complexed with Sulfonamide derivative 1, pyrophosphate, and glutamine | | Descriptor: | 5'-O-[(pyridine-3-carbonyl)sulfamoyl]adenosine, CHLORIDE ION, GLUTAMINE, ... | | Authors: | Chuenchor, W, Doukov, T.I, Gerratana, B. | | Deposit date: | 2019-03-28 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Different ways to transport ammonia in human and Mycobacterium tuberculosis NAD+synthetases.
Nat Commun, 11, 2020
|
|
5TPQ
 
 | | E. coli alkaline phosphatase D101A, D153A, R166S, E322A, K328A mutant | | Descriptor: | Alkaline phosphatase, PHOSPHATE ION, ZINC ION | | Authors: | Sunden, F, AlSadhan, I, Lyubimov, A.Y, Doukov, T, Swan, J, Herschlag, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Differential catalytic promiscuity of the alkaline phosphatase superfamily bimetallo core reveals mechanistic features underlying enzyme evolution.
J. Biol. Chem., 292, 2017
|
|
6P44
 
 | | Crystal Structure of Ketosteroid Isomerase D38N mutant from Mycobacterium hassiacum (mhKSI) bound to 3,4-dinitrophenol | | Descriptor: | 3,4-dinitrophenol, GUANIDINE, SULFATE ION, ... | | Authors: | Yabukarski, F, Doukov, T, Pinney, M, Herschlag, D. | | Deposit date: | 2019-05-25 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Parallel molecular mechanisms for enzyme temperature adaptation.
Science, 371, 2021
|
|
6P3L
 
 | | Crystal Structure of Ketosteroid Isomerase from Mycobacterium hassiacum (mhKSI) | | Descriptor: | GUANIDINE, SULFATE ION, SnoaL-like domain protein | | Authors: | Yabukarski, F, Doukov, T, Pinney, M, Herschlag, D. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Parallel molecular mechanisms for enzyme temperature adaptation.
Science, 371, 2021
|
|
6CU0
 
 | | Crystal structure of 4-1BBL/4-1BB (C121S) complex in P21 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9 | | Authors: | Aruna, B, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the human 4-1BB receptor bound to its ligand 4-1BBL reveal covalent receptor dimerization as a potential signaling amplifier.
J. Biol. Chem., 293, 2018
|
|
8SPL
 
 | | Proteinase K Multiconformer Model at 343K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SOV
 
 | | Proteinase K Multiconformer Model at 353K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SOU
 
 | | Proteinase K Multiconformer Model at 363K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SQV
 
 | | Proteinase K Multiconformer Model at 333K | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-05-04 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
