8HXG
 
 | | Crystal structure of AtHPPD-Y14116 complex | | Descriptor: | (1S,2R,3R)-2-[(S)-(2-methoxy-8-methyl-quinolin-3-yl)-oxidanyl-methyl]-6,6-dimethyl-cyclohex-4-ene-1,3-diol, (4S)-2-METHYL-2,4-PENTANEDIOL, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-04 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal structure of AtHPPD-Y14116 complex
To Be Published
|
|
8HYO
 
 | | Crystal structure of AtHPPD-Y18031 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 6-[(R)-[(2R,6R)-2,6-bis(oxidanyl)cyclohexyl]-oxidanyl-methyl]-3-(furan-2-ylmethyl)-1,5-dimethyl-quinazoline-2,4-dione, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-07 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of AtHPPD-Y18031 complex
To Be Published
|
|
8HYM
 
 | | Crystal structure of AtHPPD-Y18030 complex | | Descriptor: | 3-cyclopentyl-1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-07 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Crystal structure of AtHPPD-Y18030 complex
To Be Published
|
|
8I15
 
 | | Crystal structure of AtHPPD-YH20335 complex | | Descriptor: | 1,4-dimethyl-5-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(phenylmethyl)benzimidazol-2-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-12 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of AtHPPD-YH20335 complex
To Be Published
|
|
8I2U
 
 | | Crystal structure of AtHPPD-YH20282 complex | | Descriptor: | 2-[2-chloranyl-4-methylsulfonyl-3-(2-trimethylsilylethoxy)phenyl]carbonyl-3-oxidanyl-cyclohex-2-en-1-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Crystal structure of AtHPPD-YH20282 complex
To Be Published
|
|
8HZU
 
 | | Crystal structure of AtHPPD-XHD complex | | Descriptor: | (5R)-2-butanimidoyl-5-[(2R)-2-ethylsulfanylpropyl]-3-oxidanyl-cyclohex-2-en-1-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-09 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Crystal structure of AtHPPD-XHD complex
To Be Published
|
|
8I0Y
 
 | | Crystal structure of AtHPPD-Y191713 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, ethyl 5-methyl-6-[(2-methyl-3-oxidanylidene-1H-pyrazol-4-yl)carbonyl]-4-oxidanylidene-3-phenyl-quinazoline-2-carboxylate | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-11 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structure of AtHPPD-Y191713 complex
To Be Published
|
|
8I2S
 
 | | Crystal structure of AtHPPD-Y18979 complex | | Descriptor: | 1,5-dimethyl-3-(naphthalen-2-ylmethyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Crystal structure of AtHPPD-Y18979 complex
To Be Published
|
|
4RKF
 
 | | Drosophila melanogaster Rab3 bound to GMPPNP | | Descriptor: | MAGNESIUM ION, PENTAETHYLENE GLYCOL, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lardong, J.A, Driller, J.H, Depner, H, Weise, C, Petzoldt, A, Wahl, M.C, Sigrist, S.J, Loll, B. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of Drosophila melanogaster Rab2 and Rab3 bound to GMPPNP.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4RKE
 
 | | Drosophila melanogaster Rab2 bound to GMPPNP | | Descriptor: | GH01619p, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Lardong, J.A, Driller, J.H, Depner, H, Weise, C, Petzoldt, A, Wahl, M.C, Sigrist, S.J, Loll, B. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.0006 Å) | | Cite: | Structures of Drosophila melanogaster Rab2 and Rab3 bound to GMPPNP.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
3K3Q
 
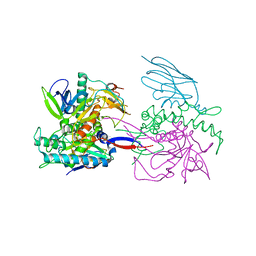 | | Crystal Structure of a Llama Antibody complexed with the C. Botulinum Neurotoxin Serotype A Catalytic Domain | | Descriptor: | Botulinum neurotoxin type A, ZINC ION, llama Aa1 VHH domain | | Authors: | Thompson, A.A, Dong, J, Marks, J.D, Stevens, R.C. | | Deposit date: | 2009-10-04 | | Release date: | 2010-02-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Single-Domain Llama Antibody Potently Inhibits the Enzymatic Activity of Botulinum Neurotoxin by Binding to the Non-Catalytic alpha-Exosite Binding Region.
J.Mol.Biol., 397, 2010
|
|
6TSH
 
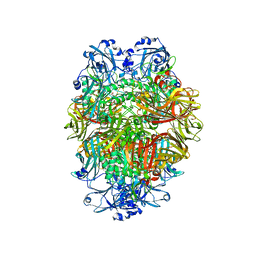 | | Beta-galactosidase in complex with deoxygalacto-nojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, Beta-galactosidase, MAGNESIUM ION | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-20 | | Release date: | 2020-01-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
6TTH
 
 | | PKM2 in complex with L-threonine | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Pyruvate kinase PKM, THREONINE | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-27 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
6TTQ
 
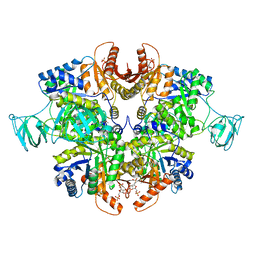 | | PKM2 in complex with Compound 10 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 1-propan-2-yl-3-pyridin-4-yl-urea, Pyruvate kinase PKM | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
8JBW
 
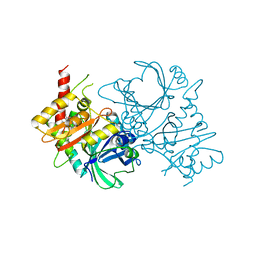 | | Crystal structure of ZtHPPD-(+)-Usnic acid complex | | Descriptor: | (9bR)-2,6-diethanoyl-8,9b-dimethyl-3,7,9-tris(oxidanyl)dibenzofuran-1-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2023-05-09 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and Development of 4-Hydroxyphenylpyruvate Dioxygenase as a Novel Crop Fungicide Target.
J.Agric.Food Chem., 71, 2023
|
|
8J8W
 
 | | Crystal structure of AtHPPD-Y19788 complex | | Descriptor: | 2,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-phenyl-quinazolin-4-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2023-05-02 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of AtHPPD-Y19788 complex
To Be Published
|
|
8J8X
 
 | | Crystal structure of AtHPPD-YH20307 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 5-(1,3-dimethyl-5-oxidanyl-pyrazol-4-yl)carbonyl-1,4-dimethyl-3-propan-2-yl-benzimidazol-2-one, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2023-05-02 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure of AtHPPD-YH20307 complex
To Be Published
|
|
8IOG
 
 | |
6TTE
 
 | | Beta-galactosidase in complex with PETG | | Descriptor: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, MAGNESIUM ION | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-27 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
6TSK
 
 | | Beta-galactosidase in complex with L-ribose | | Descriptor: | Beta-galactosidase, MAGNESIUM ION, beta-L-ribopyranose | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-20 | | Release date: | 2020-01-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
6TTF
 
 | | PKM2 in complex with Compound 5 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-hydroxynaphthalene-1-sulfonamide, Pyruvate kinase PKM | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-27 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
6TTI
 
 | | PKM2 in complex with Compound 6 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, DIMETHYL SULFOXIDE, Pyruvate kinase PKM, ... | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-27 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
8K0B
 
 | | Cryo-EM structure of TMEM63C | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Qin, Y, Yu, D, Dong, J, Dang, S. | | Deposit date: | 2023-07-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM structure of TMEM63C suggests it functions as a monomer.
Nat Commun, 14, 2023
|
|
6U07
 
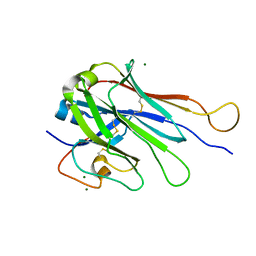 | | Computational Stabilization of T Cell Receptor Constant Domains | | Descriptor: | MAGNESIUM ION, Stabilized T cell receptor constant domain (Calpha), Stabilized T cell receptor constant domain (Cbeta) | | Authors: | Froning, K, Maguire, J, Sereno, A, Huang, F, Chang, S, Weichert, K, Frommelt, A.J, Dong, J, Wu, X, Austin, H, Conner, E.M, Fitchett, J.R, Heng, A.R, Balasubramaniam, D, Hilgers, M.T, Kuhlman, B, Demarest, S.J. | | Deposit date: | 2019-08-13 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational stabilization of T cell receptors allows pairing with antibodies to form bispecifics.
Nat Commun, 11, 2020
|
|
3T4F
 
 | | Crystal Structure of a KGE Collagen Mimetic Peptide at 1.68 A | | Descriptor: | collagen mimetic peptide | | Authors: | Fallas, J.A, Dong, J, Miller, M.D, Tao, Y.J, Hartgerink, J.D. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural insights into charge pair interactions in triple helical collagen-like proteins.
J.Biol.Chem., 287, 2012
|
|
