8HZU
 
 | | Crystal structure of AtHPPD-XHD complex | | Descriptor: | (5R)-2-butanimidoyl-5-[(2R)-2-ethylsulfanylpropyl]-3-oxidanyl-cyclohex-2-en-1-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-09 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Crystal structure of AtHPPD-XHD complex
To Be Published
|
|
8I0Y
 
 | | Crystal structure of AtHPPD-Y191713 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, ethyl 5-methyl-6-[(2-methyl-3-oxidanylidene-1H-pyrazol-4-yl)carbonyl]-4-oxidanylidene-3-phenyl-quinazoline-2-carboxylate | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-11 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structure of AtHPPD-Y191713 complex
To Be Published
|
|
8HXG
 
 | | Crystal structure of AtHPPD-Y14116 complex | | Descriptor: | (1S,2R,3R)-2-[(S)-(2-methoxy-8-methyl-quinolin-3-yl)-oxidanyl-methyl]-6,6-dimethyl-cyclohex-4-ene-1,3-diol, (4S)-2-METHYL-2,4-PENTANEDIOL, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-04 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal structure of AtHPPD-Y14116 complex
To Be Published
|
|
8HYO
 
 | | Crystal structure of AtHPPD-Y18031 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 6-[(R)-[(2R,6R)-2,6-bis(oxidanyl)cyclohexyl]-oxidanyl-methyl]-3-(furan-2-ylmethyl)-1,5-dimethyl-quinazoline-2,4-dione, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-07 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of AtHPPD-Y18031 complex
To Be Published
|
|
8I2S
 
 | | Crystal structure of AtHPPD-Y18979 complex | | Descriptor: | 1,5-dimethyl-3-(naphthalen-2-ylmethyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-15 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Crystal structure of AtHPPD-Y18979 complex
To Be Published
|
|
8HYM
 
 | | Crystal structure of AtHPPD-Y18030 complex | | Descriptor: | 3-cyclopentyl-1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-07 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Crystal structure of AtHPPD-Y18030 complex
To Be Published
|
|
8I15
 
 | | Crystal structure of AtHPPD-YH20335 complex | | Descriptor: | 1,4-dimethyl-5-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(phenylmethyl)benzimidazol-2-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-12 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of AtHPPD-YH20335 complex
To Be Published
|
|
8I2U
 
 | | Crystal structure of AtHPPD-YH20282 complex | | Descriptor: | 2-[2-chloranyl-4-methylsulfonyl-3-(2-trimethylsilylethoxy)phenyl]carbonyl-3-oxidanyl-cyclohex-2-en-1-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-15 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Crystal structure of AtHPPD-YH20282 complex
To Be Published
|
|
8HX2
 
 | | Crystal structure of AtHPPD-Y18405 complex | | Descriptor: | 3-[2-(3-chlorophenyl)ethyl]-1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexa-1,3-dien-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-03 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Crystal structure of AtHPPD-Y18405 complex
To Be Published
|
|
8HZ9
 
 | | Crystal structure of AtHPPD-Y181136 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 5-methyl-6-[(2-methyl-3-oxidanylidene-1H-pyrazol-4-yl)carbonyl]-3-propan-2-yl-1,2,3-benzotriazin-4-one, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-08 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal structure of AtHPPD-Y181136 complex
To Be Published
|
|
8HZ6
 
 | | Crystal structure of AtHPPD-QRY2089 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-prop-2-ynyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-08 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Crystal structure of AtHPPD-QRY2089 complex
To Be Published
|
|
5OBO
 
 | | Crystal structure of nitrite bound D97N mutant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, GLYCEROL, HEME C, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-28 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5OCF
 
 | | Crystal structure of nitric oxide bound to three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5OCB
 
 | | Crystal structure of nitric oxide bound D97N mutant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
6UIG
 
 | |
3QSQ
 
 | |
6E8V
 
 | | The crystal structure of bovine ultralong antibody BOV-1 | | Descriptor: | Bovine ultralong antibody BOV-1 Heavy chain, Bovine ultralong antibody BOV-1 light chain | | Authors: | Dong, J, Crowe, J.E. | | Deposit date: | 2018-07-31 | | Release date: | 2019-09-04 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Structural Diversity of Ultralong CDRH3s in Seven Bovine Antibody Heavy Chains.
Front Immunol, 10, 2019
|
|
6E9H
 
 | |
6E9U
 
 | | The crystal structure of bovine ultralong antibody BOV-7 | | Descriptor: | Bovine ultralong antibody BOV-7 heavy chain, Bovine ultralong antibody BOV-7 light chain | | Authors: | Dong, J, Crowe, J.E. | | Deposit date: | 2018-08-01 | | Release date: | 2019-05-01 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Structural Diversity of Ultralong CDRH3s in Seven Bovine Antibody Heavy Chains.
Front Immunol, 10, 2019
|
|
6E9Q
 
 | |
6E9K
 
 | |
6AGG
 
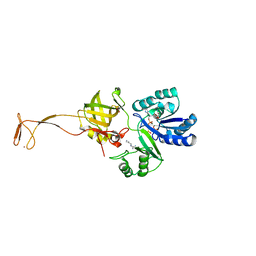 | |
6P9J
 
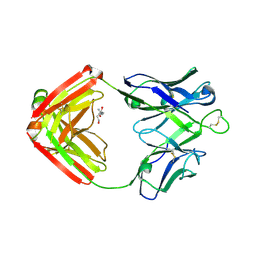 | | crystal structure of human anti staphylococcus aureus antibody STAU-229 Fab | | Descriptor: | TRIS(HYDROXYETHYL)AMINOMETHANE, human anti staphylococcus aureus antibody STAU-229 Fab heavy chain, human anti staphylococcus aureus antibody STAU-229 Fab light chain | | Authors: | Dong, J, Crowe, J.E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human V H 1-69 Gene-Encoded Human Monoclonal Antibodies against Staphylococcus aureus IsdB Use at Least Three Distinct Modes of Binding To Inhibit Bacterial Growth and Pathogenesis.
Mbio, 10, 2019
|
|
6P9I
 
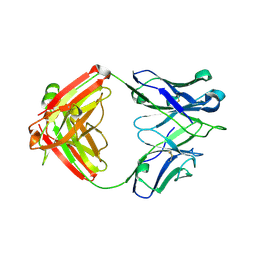 | | crystal structure of human anti staphylococcus aureus antibody STAU-399 Fab | | Descriptor: | human anti staphylococcus aureus antibody STAU-399 Fab heavy chain, human anti staphylococcus aureus antibody STAU-399 Fab light chain | | Authors: | Dong, J, Crowe, J.E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human V H 1-69 Gene-Encoded Human Monoclonal Antibodies against Staphylococcus aureus IsdB Use at Least Three Distinct Modes of Binding To Inhibit Bacterial Growth and Pathogenesis.
Mbio, 10, 2019
|
|
6P9H
 
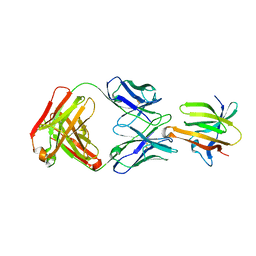 | |
