6PCK
 
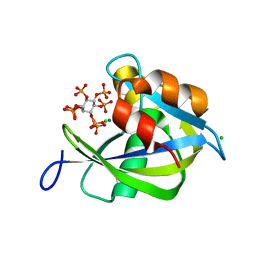 | | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 1 in complex with 1-IP7 | | Descriptor: | (1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Dollins, D.E, Neubauer, J, Dong, J, York, J.D. | | Deposit date: | 2019-06-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PCL
 
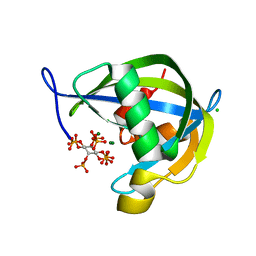 | | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 1 in complex with 5-IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Dollins, D.E, Neubauer, J, Dong, J, York, J.D. | | Deposit date: | 2019-06-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X25
 
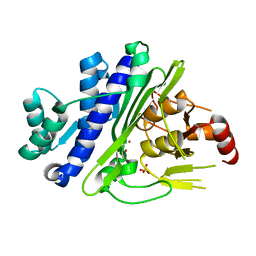 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE INPP1 IN COMPLEX GADOLINIUM AFTER ADDITION OF INOSITOL 1,3,4-TRISPHOSPHATE AND LITHIUM AT 3.2 ANGSTROM RESOLUTION | | Descriptor: | GADOLINIUM ATOM, Inositol polyphosphate 1-phosphatase, SULFATE ION | | Authors: | Dollins, D.E, Endo-Streeter, S, Ren, Y, York, J.D. | | Deposit date: | 2020-05-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
6WRO
 
 | |
7KIR
 
 | | Crystal structure of inositol polyphosphate 1-phosphatase (INPP1) D54A mutant in complex with inositol (1,4)-bisphosphate | | Descriptor: | CALCIUM ION, D-MYO-INOSITOL-1,4-BISPHOSPHATE, Inositol polyphosphate 1-phosphatase | | Authors: | Dollins, D.E, Xiong, J.-P, Ren, Y, York, J.D. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
1YT0
 
 | | Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N(DELTA)41 APO CRYSTAL SOAKED WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Dollins, D.E, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Unliganded GRP94, the Endoplasmic Reticulum Hsp90: BASIS FOR NUCLEOTIDE-INDUCED CONFORMATIONAL CHANGE
J.Biol.Chem., 280, 2005
|
|
1YSZ
 
 | | Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N(DELTA)41 APO CRYSTAL SOAKED WITH NECA | | Descriptor: | Endoplasmin, N-ETHYL-5'-CARBOXAMIDO ADENOSINE, TETRAETHYLENE GLYCOL | | Authors: | Dollins, D.E, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of Unliganded GRP94, the Endoplasmic Reticulum Hsp90: BASIS FOR NUCLEOTIDE-INDUCED CONFORMATIONAL CHANGE
J.Biol.Chem., 280, 2005
|
|
1YT1
 
 | | Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N(DELTA)41 APO CRYSTAL | | Descriptor: | Endoplasmin, PENTAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL | | Authors: | Dollins, D.E, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Unliganded GRP94, the Endoplasmic Reticulum Hsp90: BASIS FOR NUCLEOTIDE-INDUCED CONFORMATIONAL CHANGE
J.Biol.Chem., 280, 2005
|
|
1YT2
 
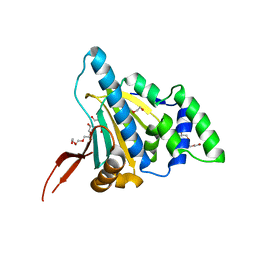 | | Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N APO CRYSTAL | | Descriptor: | Endoplasmin, TETRAETHYLENE GLYCOL | | Authors: | Dollins, D.E, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of Unliganded GRP94, the Endoplasmic Reticulum Hsp90: BASIS FOR NUCLEOTIDE-INDUCED CONFORMATIONAL CHANGE
J.Biol.Chem., 280, 2005
|
|
2O1T
 
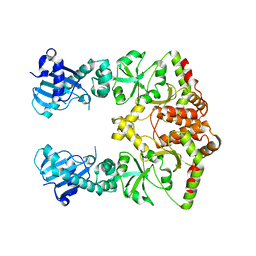 | |
2O1U
 
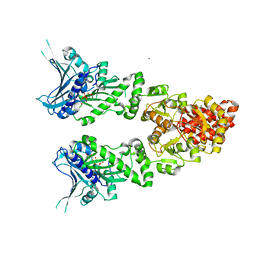 | | Structure of full length GRP94 with AMP-PNP bound | | Descriptor: | Endoplasmin, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Dollins, D.E, Warren, J.J, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2006-11-29 | | Release date: | 2007-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of GRP94-Nucleotide Complexes Reveal Mechanistic Differences between the hsp90 Chaperones.
Mol.Cell, 28, 2007
|
|
2O1V
 
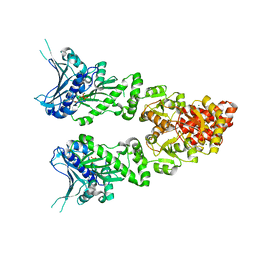 | | Structure of full length GRP94 with ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Endoplasmin, MAGNESIUM ION | | Authors: | Dollins, D.E, Warren, J.J, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2006-11-29 | | Release date: | 2007-10-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of GRP94-Nucleotide Complexes Reveal Mechanistic Differences between the hsp90 Chaperones.
Mol.Cell, 28, 2007
|
|
2O1W
 
 | |
1R4I
 
 | | Crystal Structure of Androgen Receptor DNA-Binding Domain Bound to a Direct Repeat Response Element | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*TP*CP*AP*AP*GP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*CP*TP*TP*GP*AP*TP*GP*TP*TP*CP*TP*GP*G)-3', Androgen receptor, ... | | Authors: | Shaffer, P.L, Jivan, A, Dollins, D.E, Claessens, F, Gewirth, D.T. | | Deposit date: | 2003-10-06 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of androgen receptor binding to selective androgen response elements.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
7KIO
 
 | | Crystal structure of inositol polyphosphate 1-phosphatase (INPP1) D54A mutant | | Descriptor: | CALCIUM ION, Inositol polyphosphate 1-phosphatase, SULFATE ION | | Authors: | Xiong, J.-P, Dollins, D.E, Ren, Y, York, J.D. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
4HNY
 
 | | Apo N-terminal acetyltransferase complex A | | Descriptor: | GLYCEROL, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1, ... | | Authors: | Neubauer, J.L, Immormino, R.M, Dollins, D.E, Endo-Streeter, S.T, Pemble IV, C.W, York, J.D. | | Deposit date: | 2012-10-21 | | Release date: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
To be Published
|
|
4HNW
 
 | | The NatA Acetyltransferase Complex Bound To Inositol Hexakisphosphate | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1, ... | | Authors: | Neubauer, J.L, Immormino, R.M, Dollins, D.E, Endo-Streeter, S.T, Pemble IV, C.W, York, J.D. | | Deposit date: | 2012-10-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
To be Published
|
|
4HNX
 
 | | The NatA Acetyltransferase Complex Bound To ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1 | | Authors: | Neubauer, J.L, Immormino, R.M, Dollins, D.E, Endo-Streeter, S.T, Pemble IV, C.W, York, J.D. | | Deposit date: | 2012-10-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
To be Published
|
|
1TBW
 
 | | Ligand Induced Conformational Shift in the N-terminal Domain of GRP94, Open Conformation | | Descriptor: | ADENOSINE MONOPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Gewirth, D.T, Immormino, R.M, Dollins, D.E, Shaffer, P.L, Walker, M.A, Soldano, K.L. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand-induced Conformational Shift in the N-terminal Domain of GRP94, an Hsp90 Chaperone.
J.Biol.Chem., 279, 2004
|
|
1TC0
 
 | | Ligand Induced Conformational Shifts in the N-terminal Domain of GRP94, Open Conformation Complexed with the physiological partner ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Gewirth, D.T, Immormino, R.M, Dollins, D.E, Shaffer, P.L, Walker, M.A, Soldano, K.L. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-induced Conformational Shift in the N-terminal Domain of GRP94, an Hsp90 Chaperone.
J.Biol.Chem., 279, 2004
|
|
1TC6
 
 | | Ligand Induced Conformational Shift in the N-terminal Domain of GRP94, Open Conformation ADP-Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Gewirth, D.T, Immormino, R.M, Dollins, D.E, Shaffer, P.L, Walker, M.A, Soldano, K.L. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Ligand-induced Conformational Shift in the N-terminal Domain of GRP94, an Hsp90 Chaperone.
J.Biol.Chem., 279, 2004
|
|
