6Q26
 
 | |
5KZD
 
 | | N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus with bound sialic acid alditol | | Descriptor: | (2~{S},4~{S},5~{R},6~{R},7~{S},8~{R})-5-acetamido-2,4,6,7,8,9-hexakis(oxidanyl)nonanoic acid, N-acetylneuraminate lyase | | Authors: | North, R.A, Watson, A.J.A, Pearce, F.G, Muscroft-Taylor, A.C, Friemann, R, Fairbanks, A.J, Dobson, R.C.J. | | Deposit date: | 2016-07-25 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Structure and inhibition of N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus.
FEBS Lett., 590, 2016
|
|
6H4E
 
 | | Proteus mirabilis N-acetylneuraminate lyase | | Descriptor: | Putative N-acetylneuraminate lyase, SULFATE ION | | Authors: | North, R.A, Garcia-Bonete, M.J, Goyal, P, Katona, G, Dobson, R.C.J, Friemann, R. | | Deposit date: | 2018-07-21 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | The structure of Proteus mirabilis N-acetylneuraminate lyase reveals an intermolecular disulphide bond
To Be Published
|
|
4IK0
 
 | | Crystal structure of diaminopimelate epimerase Y268A mutant from Escherichia coli | | Descriptor: | Diaminopimelate epimerase, IODIDE ION | | Authors: | Hor, L, Dobson, R.C.J, Hutton, C.A, Perugini, M.A. | | Deposit date: | 2012-12-24 | | Release date: | 2013-02-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dimerization of bacterial diaminopimelate epimerase is essential for catalysis
J.Biol.Chem., 288, 2013
|
|
4IJZ
 
 | | Crystal structure of diaminopimelate epimerase from Escherichia coli | | Descriptor: | Diaminopimelate epimerase, NITRATE ION | | Authors: | Hor, L, Dobson, R.C.J, Hutton, C.A, Perugini, M.A. | | Deposit date: | 2012-12-24 | | Release date: | 2013-02-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dimerization of bacterial diaminopimelate epimerase is essential for catalysis
J.Biol.Chem., 288, 2013
|
|
2OJP
 
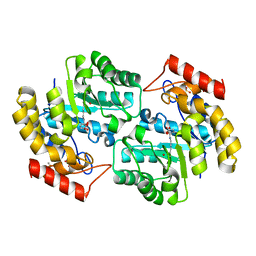 | | The crystal structure of a dimeric mutant of Dihydrodipicolinate synthase from E.coli- DHDPS-L197Y | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL | | Authors: | Griffin, M.D.W, Dobson, R.C.J, Antonio, L, Perugini, M.A, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2007-01-13 | | Release date: | 2008-01-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of quaternary structure in a homotetrameric enzyme.
J.Mol.Biol., 380, 2008
|
|
7RUM
 
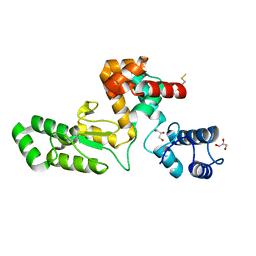 | | Endolysin from Escherichia coli O157:H7 phage FTEbC1, LysT84 | | Descriptor: | Endolysin, GLYCEROL | | Authors: | Love, M.J, Billington, C, Dobson, R.C.J. | | Deposit date: | 2021-08-17 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The structure and function of modular Escherichia coli O157:H7 bacteriophage FTBEc1 endolysin, LysT84: defining a new endolysin catalytic subfamily.
Biochem.J., 479, 2022
|
|
8THI
 
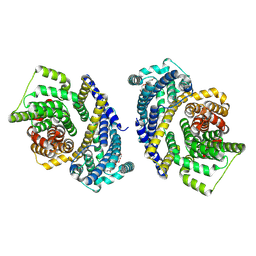 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (parallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, SODIUM ION, Sialic acid TRAP transporter permease protein SiaT | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
8THJ
 
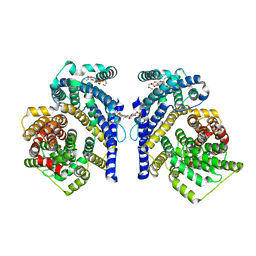 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (antiparallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
4C1L
 
 | | Crystal structure of pyrococcus furiosus 3-deoxy-D-arabino- heptulosonate 7-phosphate synthase I181D interface mutant | | Descriptor: | 2-dehydro-3-deoxyphosphoheptonate aldolase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Nazmi, A.R, Schofield, L.R, Dobson, R.C.J, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Destabilization of the homotetrameric assembly of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from the hyperthermophile Pyrococcus furiosus enhances enzymatic activity.
J. Mol. Biol., 426, 2014
|
|
6Q27
 
 | | N-acetylmannosamine kinase with N-acetylmannosamine from Staphylococcus aureus | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-mannopyranose, Glucokinase | | Authors: | Coombes, D, Horne, C.R, Davies, J.S, Dobson, R.C.J. | | Deposit date: | 2019-08-07 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The basis for non-canonical ROK family function in theN-acetylmannosamine kinase from the pathogenStaphylococcus aureus.
J.Biol.Chem., 295, 2020
|
|
4OY5
 
 | | 0.89 Angstrom resolution crystal structure of (Gly-Pro-Hyp)10 | | Descriptor: | Collagen | | Authors: | Suzuki, H, Mahapatra, D, Steel, P.J, Dyer, J, Dobson, R.C.J, Gerrard, J.A, Valery, C. | | Deposit date: | 2014-02-10 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Sub-angstrom structure of the collagen model peptide (GPO)10 shows a hydrated triple helix with pitch variation and two proline ring conformations
To Be Published
|
|
6Q28
 
 | |
8T9T
 
 | |
8TE9
 
 | |
5DPT
 
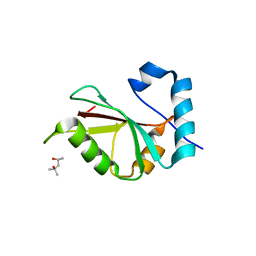 | | Crystal structure of PLEKHM1 LIR-fused human GABARAPL1_2-117 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Pleckstrin homology domain-containing family M member 1, Gamma-aminobutyric acid receptor-associated protein-like 1,Gamma-aminobutyric acid receptor-associated protein-like 1 | | Authors: | Ravichandran, A.C, Suzuki, H, Dobson, R.C.J. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional analysis of the GABARAP interaction motif (GIM).
EMBO Rep., 18, 2017
|
|
5DPR
 
 | |
5DPS
 
 | |
5DPW
 
 | |
4C1K
 
 | | Crystal structure of pyrococcus furiosus 3-deoxy-D-arabino- heptulosonate 7-phosphate synthase | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOHEPTONATE ALDOLASE, CADMIUM ION, CARBONATE ION, ... | | Authors: | Nazmi, A.R, Schofield, L.R, Dobson, R.C.J, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Destabilization of the Homotetrameric Assembly of 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase from the Hyperthermophile Pyrococcus Furiosus Enhances Enzymatic Activity
J.Mol.Biol., 426, 2014
|
|
8EDQ
 
 | | E. coli pyruvate kinase (PykF) I264F | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pyruvate kinase, SULFATE ION | | Authors: | Donovan, K.A, Coombes, D, Dobson, R.C.J, Cooper, T.F. | | Deposit date: | 2022-09-05 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Beneficial mutations occurring in E. coli pyruvate kinase afford new allosteric mechanisms leading to faster resumption of growth
To Be Published
|
|
8EDT
 
 | |
8EDS
 
 | |
8EDR
 
 | |
8EQ0
 
 | |
