8IGI
 
 | | Crystal structure of HP1526 (XthA)- a base excision DNA repair protein in Helicobacter pylori | | Descriptor: | 1,3-BUTANEDIOL, Exodeoxyribonuclease (LexA), MANGANESE (II) ION | | Authors: | Dinh, T.T, Dao, O, Lee, K.H. | | Deposit date: | 2023-02-20 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the apurinic/apyrimidinic endonuclease XthA (HP1526 protein) from Helicobacter pylori.
Biochem.Biophys.Res.Commun., 663, 2023
|
|
8GQQ
 
 | |
6XNB
 
 | |
8D3S
 
 | | HIV-1 Integrase Catalytic Core Domain F185H Mutant Complexed with BKC-110 | | Descriptor: | (2S)-tert-butoxy{4-(4-chlorophenyl)-2,6-dimethyl-1-[(1-methyl-1H-pyrazol-4-yl)methyl]-1H-pyrrolo[2,3-b]pyridin-5-yl}acetic acid, Integrase | | Authors: | Dinh, T, Kvaratskhelia, M. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structural and mechanistic bases for the viral resistance to allosteric HIV-1 integrase inhibitor pirmitegravir.
Biorxiv, 2024
|
|
7UTC
 
 | |
7UUT
 
 | | Ternary complex crystal structure of secondary alcohol dehydrogenases from the Thermoanaerobacter ethanolicus mutants C295A and I86A provides better understanding of catalytic mechanism | | Descriptor: | (2R)-pentan-2-ol, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Dinh, T, Phillips, R, Rahn, K. | | Deposit date: | 2022-04-28 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystallographic snapshots of ternary complexes of thermophilic secondary alcohol dehydrogenase from Thermoanaerobacter pseudoethanolicus reveal the dynamics of ligand exchange and the proton relay network.
Proteins, 90, 2022
|
|
8S9Q
 
 | |
8T5B
 
 | |
8T5A
 
 | |
8T52
 
 | |
2D47
 
 | | MOLECULAR STRUCTURE OF A COMPLETE TURN OF A-DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*CP*CP*GP*CP*GP*GP*GP*GP*G)-3'), SPERMINE | | Authors: | Verdaguer, N, Aymami, J, Fernandez-Forner, D, Fita, I, Coll, M, Huynh-Dinh, T, Igolen, J, Subirana, J.A. | | Deposit date: | 1991-10-02 | | Release date: | 1991-10-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of a complete turn of A-DNA.
J.Mol.Biol., 221, 1991
|
|
1ESK
 
 | | SOLUTION STRUCTURE OF NCP7 FROM HIV-1 | | Descriptor: | GAG POLYPROTEIN, ZINC ION | | Authors: | Morellet, N, Demene, H, Teilleux, V, Huynh-Dinh, T, de Rocquigny, H, Fournie-Zaluski, M.-C, Roques, B.P. | | Deposit date: | 2000-04-10 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of (12-53)NCp7 of HIV-1
To be Published
|
|
249D
 
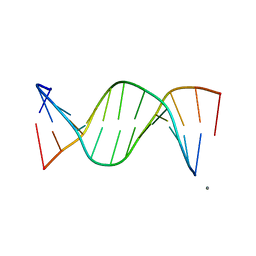 | | STRUCTURAL COMPARISON BETWEEN THE D(CTAG) SEQUENCE IN OLIGONUCLEOTIDES AND TRP AND MET REPRESSOR-OPERATOR COMPLEXES | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*CP*TP*CP*TP*AP*GP*AP*GP*CP*G)-3') | | Authors: | Urpi, L, Tereshko, V, Malinina, L, Huynh-Dinh, T, Subirana, J.A. | | Deposit date: | 1996-02-22 | | Release date: | 1996-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural comparison between the d(CTAG) sequence in oligonucleotides and trp and met repressor-operator complexes.
Nat.Struct.Biol., 3, 1996
|
|
250D
 
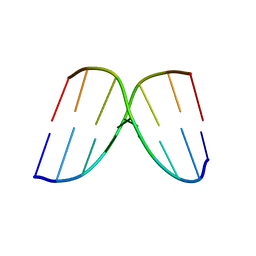 | | STRUCTURAL COMPARISON BETWEEN THE D(CTAG) SEQUENCE IN OLIGONUCLEOTIDES AND TRP AND MET REPRESSOR-OPERATOR COMPLEXES | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3') | | Authors: | Urpi, L, Tereshko, V, Malinina, L, Huynh-Dinh, T, Subirana, J.A. | | Deposit date: | 1996-02-22 | | Release date: | 1996-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural comparison between the d(CTAG) sequence in oligonucleotides and trp and met repressor-operator complexes.
Nat.Struct.Biol., 3, 1996
|
|
192D
 
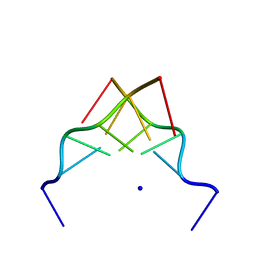 | | RECOMBINATION-LIKE STRUCTURE OF D(CCGCGG) | | Descriptor: | DNA (5'-D(*CP*CP*GP*CP*GP*G)-3'), SODIUM ION | | Authors: | Malinina, L, Urpi, L, Salas, X, Huynh-Dinh, T, Subirana, J.A. | | Deposit date: | 1994-09-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Recombination-like structure of d(CCGCGG).
J.Mol.Biol., 243, 1994
|
|
1BJ6
 
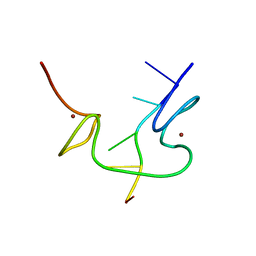 | | 1H NMR OF (12-53) NCP7/D(ACGCC) COMPLEX, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*C)-3'), NUCLEOCAPSID PROTEIN 7, ZINC ION | | Authors: | Demene, H, Morellet, N, Teilleux, V, Huynh-Dinh, T, De Rocquigny, H, Fournie-Zaluski, M.C, Roques, B.P. | | Deposit date: | 1998-07-03 | | Release date: | 1999-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC).
J.Mol.Biol., 283, 1998
|
|
446D
 
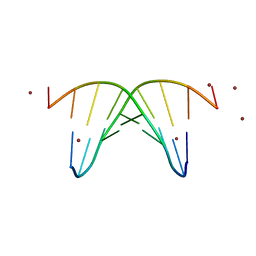 | | STRUCTURE OF THE OLIGONUCLEOTIDE D(CGTATATACG) AS A SITE SPECIFIC COMPLEX WITH NICKEL IONS | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*TP*AP*TP*AP*CP*G)-3'), NICKEL (II) ION | | Authors: | Abrescia, N.G.A, Malinina, L, Gonzaga, L.F, Huynh-Dinh, T, Neidle, S, Subirana, J.A. | | Deposit date: | 1999-01-15 | | Release date: | 1999-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the oligonucleotide d(CGTATATACG) as a site-specific complex with nickel ions.
Nucleic Acids Res., 27, 1999
|
|
1U3Q
 
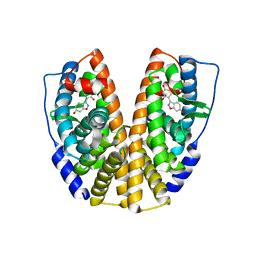 | | Crystal Structure of Estrogen Receptor beta complexed with CL-272 | | Descriptor: | 4-(6-HYDROXY-BENZO[D]ISOXAZOL-3-YL)BENZENE-1,3-DIOL, Estrogen receptor beta | | Authors: | Malamas, M.S, Manas, E.S, McDevitt, R.E, Gunawan, I, Xu, Z.B, Collini, M.D, Miller, C.P, Dinh, T, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2004-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of aryl diphenolic azoles as potent and selective estrogen receptor-beta ligands.
J.Med.Chem., 47, 2004
|
|
1U3R
 
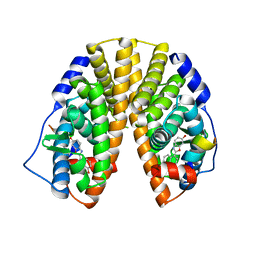 | | Crystal Structure of Estrogen Receptor beta complexed with WAY-338 | | Descriptor: | 2-(5-HYDROXY-NAPHTHALEN-1-YL)-1,3-BENZOOXAZOL-6-OL, Estrogen receptor beta, steroid receptor coactivator-1 | | Authors: | Malamas, M.S, Manas, E.S, McDevitt, R.E, Gunawan, I, Xu, Z.B, Collini, M.D, Miller, C.P, Dinh, T, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2004-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Design and synthesis of aryl diphenolic azoles as potent and selective estrogen receptor-beta ligands.
J.Med.Chem., 47, 2004
|
|
1U3S
 
 | | Crystal Structure of Estrogen Receptor beta complexed with WAY-797 | | Descriptor: | 3-(6-HYDROXY-NAPHTHALEN-2-YL)-BENZO[D]ISOOXAZOL-6-OL, Estrogen receptor beta, steroid receptor coactivator-1 | | Authors: | Malamas, M.S, Manas, E.S, McDevitt, R.E, Gunawan, I, Xu, Z.B, Collini, M.D, Miller, C.P, Dinh, T, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2004-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and synthesis of aryl diphenolic azoles as potent and selective estrogen receptor-beta ligands.
J.Med.Chem., 47, 2004
|
|
1GQU
 
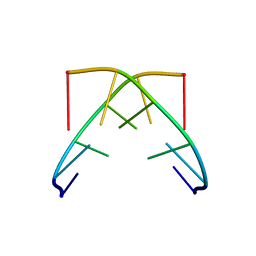 | |
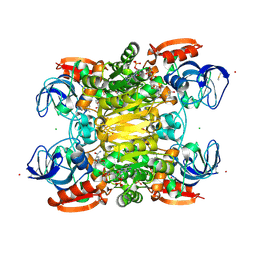
7UX4
 
 | |
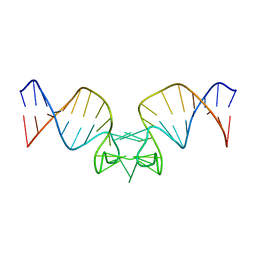
1JU0
 
 | |
1JTJ
 
 | | Solution structure of HIV-1Lai mutated SL1 hairpin | | Descriptor: | HIV-1Lai SL1 | | Authors: | Kieken, F, Arnoult, E, Barbault, F, Paquet, F, Huynh-Dinh, T, Paoletti, J, Genest, D, Lancelot, G. | | Deposit date: | 2001-08-21 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HIV-1(Lai) genomic RNA: combined used of NMR and molecular dynamics simulation for studying
the structure and internal dynamics of a mutated SL1 hairpin.
EUR.BIOPHYS.J., 31, 2002
|
|
1JUA
 
 | |
