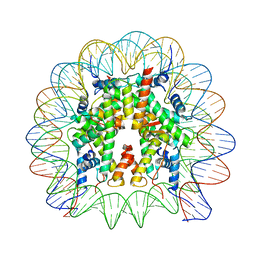
8JBX
 
 | |
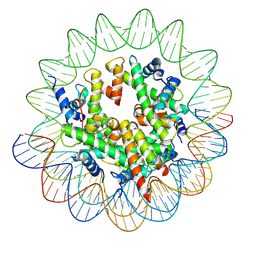
8JCC
 
 | |
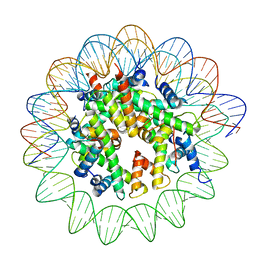
8JCD
 
 | |
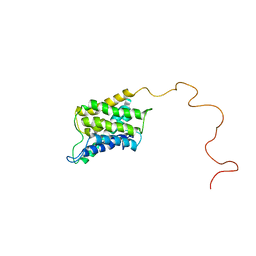
6POJ
 
 | |
1I2Z
 
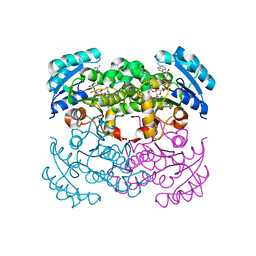 | | E. COLI ENOYL REDUCTASE IN COMPLEX WITH NAD AND BRL-12654 | | Descriptor: | 4-(2-THIENYL)-1-(4-METHYLBENZYL)-1H-IMIDAZOLE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Heerding, D.A, Miller, W.H, Payne, D.J, Janson, C.A, Qiu, X. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 1,4-Disubstituted imidazoles are potential antibacterial agents functioning as inhibitors of enoyl acyl carrier protein
reductase (FabI).
Bioorg.Med.Chem.Lett., 11, 2001
|
|
8JNJ
 
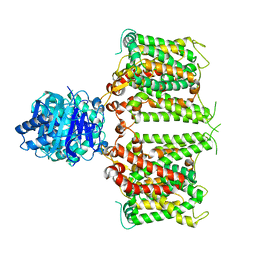 | | Structure of R932A/K1147A/H1148A mutant AE2 | | Descriptor: | Anion exchange protein 2 | | Authors: | Yin, Y.X, Ding, D. | | Deposit date: | 2023-06-06 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional insights into the lipid regulation of human anion exchanger 2.
Nat Commun, 15, 2024
|
|
8JNI
 
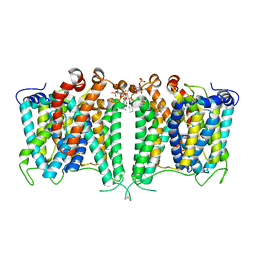 | | Structure of AE2 in complex with PIP2 | | Descriptor: | Anion exchange protein 2, CHLORIDE ION, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate | | Authors: | Yin, Y.X, Ding, D. | | Deposit date: | 2023-06-06 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into the lipid regulation of human anion exchanger 2.
Nat Commun, 15, 2024
|
|
7Y1K
 
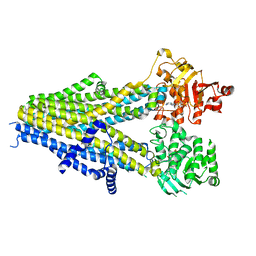 | | Structure of SUR2A in complex with Mg-ATP, Mg-ADP and repaglinide in the inward-facing conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 9, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1L
 
 | | Structure of SUR2B in complex with Mg-ATP and repaglinide in the inward-facing conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Isoform SUR2B of ATP-binding cassette sub-family C member 9, MAGNESIUM ION, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1J
 
 | | Structure of SUR2A in complex with Mg-ATP and repaglinide in the inward-facing conformation. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 9, MAGNESIUM ION, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1M
 
 | | Structure of SUR2B in complex with Mg-ATP, Mg-ADP, and repaglinide in the inward-facing conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Isoform SUR2B of ATP-binding cassette sub-family C member 9, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1N
 
 | | Structure of SUR2B in complex with Mg-ATP, Mg-ADP, and repaglinide in the partially occluded state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Isoform SUR2B of ATP-binding cassette sub-family C member 9, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7VLT
 
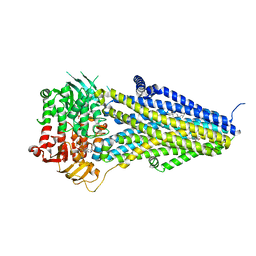 | | Structure of SUR2B in complex with Mg-ATP/ADP and levcromakalim | | Descriptor: | (3S,4R)-2,2-dimethyl-3-oxidanyl-4-(2-oxidanylidenepyrrolidin-1-yl)-3,4-dihydrochromene-6-carbonitrile, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
7VLU
 
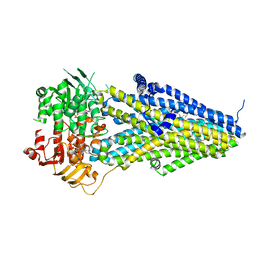 | | Structure of SUR2A in complex with Mg-ATP/ADP and P1075 | | Descriptor: | 1-cyano-2-(2-methylbutan-2-yl)-3-pyridin-3-yl-guanidine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
7VLR
 
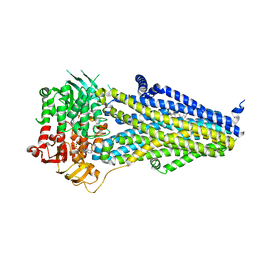 | | Structure of SUR2B in complex with Mg-ATP/ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
7VLS
 
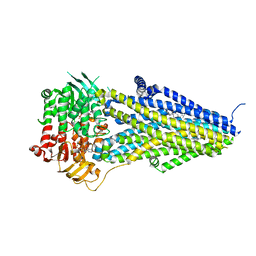 | | Structure of SUR2B in complex with MgATP/ADP and P1075 | | Descriptor: | 1-cyano-2-(2-methylbutan-2-yl)-3-pyridin-3-yl-guanidine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
6JB3
 
 | | Structure of SUR1 subunit bound with repaglinide | | Descriptor: | ATP-binding cassette sub-family C member 8 isoform X2, Digitonin, Repaglinide | | Authors: | Chen, L, Ding, D, Wang, M, Wu, J.-X, Kang, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | The Structural Basis for the Binding of Repaglinide to the Pancreatic KATPChannel.
Cell Rep, 27, 2019
|
|
6JB1
 
 | | Structure of pancreatic ATP-sensitive potassium channel bound with repaglinide and ATPgammaS at 3.3A resolution | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ATP-binding cassette sub-family C member 8 isoform X2, ATP-sensitive inward rectifier potassium channel 11, ... | | Authors: | Chen, L, Ding, D, Wang, M, Wu, J.-X, Kang, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The Structural Basis for the Binding of Repaglinide to the Pancreatic KATPChannel.
Cell Rep, 27, 2019
|
|
1G5S
 
 | | CRYSTAL STRUCTURE OF HUMAN CYCLIN DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH THE INHIBITOR H717 | | Descriptor: | 2-[TRANS-(4-AMINOCYCLOHEXYL)AMINO]-6-(BENZYL-AMINO)-9-CYCLOPENTYLPURINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Dreyer, M.K, Borcherding, D.R, Dumont, J.A, Peet, N.P, Tsay, J.T, Wright, P.S, Bitonti, A.J, Shen, J, Kim, S.-H. | | Deposit date: | 2000-11-02 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of human cyclin-dependent kinase 2 in complex with the adenine-derived inhibitor H717.
J.Med.Chem., 44, 2001
|
|
5EZR
 
 | | Crystal Structure of PVX_084705 bound to compound | | Descriptor: | CHLORIDE ION, N-[5-(3-{2-[(cyclopropylmethyl)amino]pyrimidin-4-yl}-7-[(dimethylamino)methyl]-6-methylimidazo[1,2-a]pyridin-2-yl)-2-fluorophenyl]methanesulfonamide, cGMP-dependent protein kinase, ... | | Authors: | El Bakkouri, M, Amani, M, Walker, J.R, Osborne, S, Large, J.M, Birchall, K, Bouloc, N, Smiljanic-Hurley, E, Wheldon, M, Harding, D.J, Merritt, A.T, Ansell, K.H, Coombs, P.J, Kettleborough, C.A, Stewart, B.L, Bowyer, P.W, Gutteridge, W.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Baker, D.A, Hui, R, Loppnau, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-26 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PVX_084705 bound to compound
To Be Published
|
|
3QCY
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 4-[2-Amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-N-phenyl-2-morpholinecarboxamide | | Descriptor: | (2S)-4-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-N-phenylmorpholine-2-carboxamide, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QCX
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-{2-Amino-6-[(3R)-3-methyl-4-morpholinyl]-4-pyrimidinyl}-1H-indazol-3-amine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-{2-amino-6-[(3R)-3-methylmorpholin-4-yl]pyrimidin-4-yl}-2H-indazol-3-amine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QCQ
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-(3-Amino-1H-indazol-6-yl)-N4-ethyl-2,4-pyrimidinediamine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-(3-amino-2H-indazol-6-yl)-N~4~-ethylpyrimidine-2,4-diamine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QD3
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 1,1-Dimethylethyl {(3R,6S)-1-[2-amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-6-methyl-3-piperidinyl}carbamate | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QD4
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 1,1-Dimethylethyl{(3R,5R)-1-[2-amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-5-methyl-3-piperidinyl}carbamate | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, SULFATE ION, tert-butyl {(3R,5R)-1-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-5-methylpiperidin-3-yl}carbamate | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
