7RSL
 
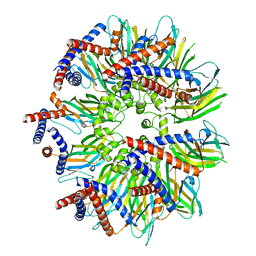 | | Seipin forms a flexible cage at lipid droplet formation sites | | 分子名称: | Seipin | | 著者 | Arlt, H, Sui, X, Folger, B, Adams, C, Chen, X, Remme, R, Hamprecht, F.A, DiMaio, F, Liao, M, Goodman, J.M, Farese Jr, R.V, Walther, T.C. | | 登録日 | 2021-08-11 | | 公開日 | 2022-02-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Seipin forms a flexible cage at lipid droplet formation sites.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7K3H
 
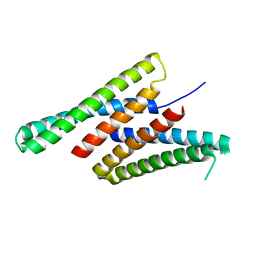 | | Crystal structure of deep network hallucinated protein 0217 | | 分子名称: | Network hallucinated protein 0217 | | 著者 | Pellock, S.J, Anishchenko, I, Chidyausiku, T.M, Bera, A.K, DiMaio, F, Baker, D. | | 登録日 | 2020-09-11 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | De novo protein design by deep network hallucination.
Nature, 600, 2021
|
|
5J0L
 
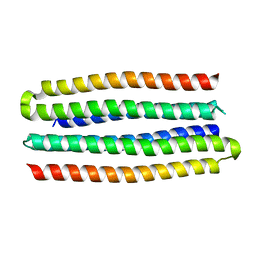 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | 分子名称: | designed protein 3L6HC2_2 | | 著者 | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | 登録日 | 2016-03-28 | | 公開日 | 2016-07-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
7JZV
 
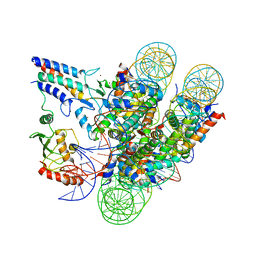 | | Cryo-EM structure of the BRCA1-UbcH5c/BARD1 E3-E2 module bound to a nucleosome | | 分子名称: | BRCA1,Ubiquitin-conjugating enzyme E2 D3, BRCA1-associated RING domain protein 1, Histone H2A type 2-A, ... | | 著者 | Witus, S.R, Burrell, A.L, Hansen, J.M, Farrell, D.P, Dimaio, F, Kollman, J.M, Klevit, R.E. | | 登録日 | 2020-09-02 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7O1Q
 
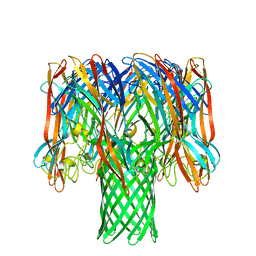 | | Amyloid beta oligomer displayed on the alpha hemolysin scaffold | | 分子名称: | Alpha-hemolysin hybridized Abeta | | 著者 | Wu, J, Blum, T.B, Farrell, D.P, DiMaio, F, Abrahams, J.P, Luo, J. | | 登録日 | 2021-03-30 | | 公開日 | 2021-04-14 | | 最終更新日 | 2021-08-18 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-electron Microscopy Imaging of Alzheimer's Amyloid-beta 42 Oligomer Displayed on a Functionally and Structurally Relevant Scaffold.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8UB3
 
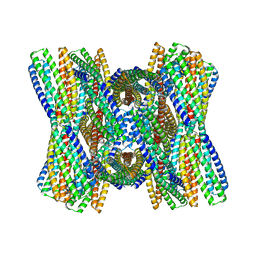 | | DpHF7 filament | | 分子名称: | DpHF7 filament | | 著者 | Lynch, E.M, Farrell, D, Shen, H, Kollman, J.M, DiMaio, F, Baker, D. | | 登録日 | 2023-09-22 | | 公開日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 2024
|
|
5J0I
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | 分子名称: | Designed protein 2L6HC3_12 | | 著者 | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | 登録日 | 2016-03-28 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J0K
 
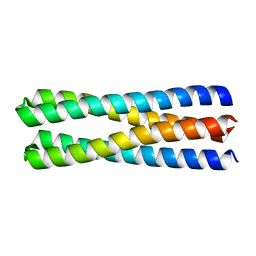 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | 分子名称: | designed protein 2L4HC2_23 | | 著者 | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | 登録日 | 2016-03-28 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J10
 
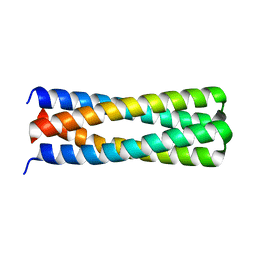 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | 分子名称: | peptide design 2L4HC2_24 | | 著者 | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | 登録日 | 2016-03-28 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J0H
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | 分子名称: | Design construct 2L6HC3_13 | | 著者 | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | 登録日 | 2016-03-28 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
3SQF
 
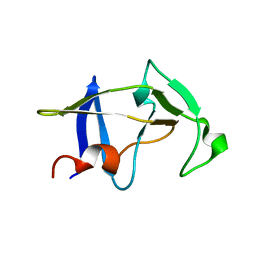 | | Crystal structure of monomeric M-PMV retroviral protease | | 分子名称: | Protease | | 著者 | Jaskolski, M, Kazmierczyk, M, Gilski, M, Krzywda, S, Pichova, I, Zabranska, H, Khatib, F, DiMaio, F, Cooper, S, Thompson, J, Popovic, Z, Baker, D, Group, Foldit Contenders | | 登録日 | 2011-07-05 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6324 Å) | | 主引用文献 | Crystal structure of a monomeric retroviral protease solved by protein folding game players.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8DIT
 
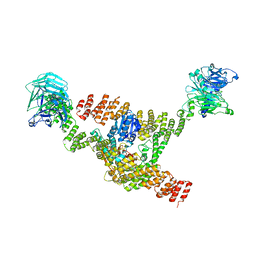 | | Cryo-EM structure of a HOPS core complex containing Vps33, Vps16, and Vps18 | | 分子名称: | Vacuolar protein sorting-associated protein 16, Vacuolar protein sorting-associated protein 18, Vacuolar protein sorting-associated protein 33 | | 著者 | Port, S.A, Farrell, P.D, Jeffrey, P.D, DiMaio, F, Hughson, F.M. | | 登録日 | 2022-06-29 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Cryo-EM structure of the HOPS core complex and its implication for SNARE assembly
To Be Published
|
|
6Q04
 
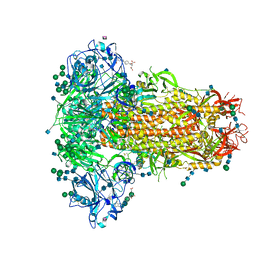 | | MERS-CoV S structure in complex with 5-N-acetyl neuraminic acid | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | 著者 | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2019-08-01 | | 公開日 | 2019-12-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q06
 
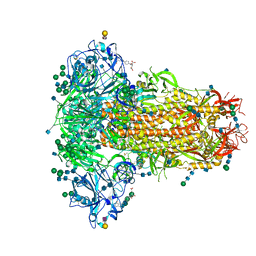 | | MERS-CoV S structure in complex with 2,3-sialyl-N-acetyl-lactosamine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | 著者 | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2019-08-01 | | 公開日 | 2019-12-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q05
 
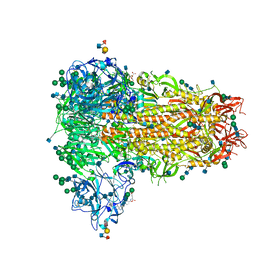 | | MERS-CoV S structure in complex with sialyl-lewisX | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | 著者 | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2019-08-01 | | 公開日 | 2019-12-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q07
 
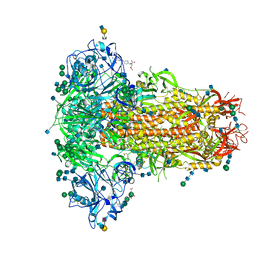 | | MERS-CoV S structure in complex with 2,6-sialyl-N-acetyl-lactosamine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | 著者 | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2019-08-01 | | 公開日 | 2019-12-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7RMC
 
 | | Yeast CTP Synthase (Ura7) filament bound to CTP at low pH | | 分子名称: | CTP synthase 1, CYTIDINE-5'-TRIPHOSPHATE | | 著者 | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | 登録日 | 2021-07-27 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RMO
 
 | | Yeast CTP Synthase (Ura7) Bundle bound to Products at low pH | | 分子名称: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE | | 著者 | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | 登録日 | 2021-07-27 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RNR
 
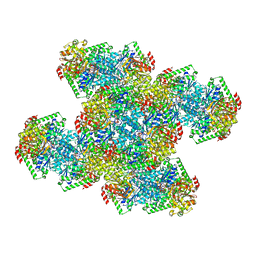 | | Yeast CTP Synthase (Ura8) Bundle Bound to Substrates at Low pH | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, MAGNESIUM ION, ... | | 著者 | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | 登録日 | 2021-07-29 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
5W7G
 
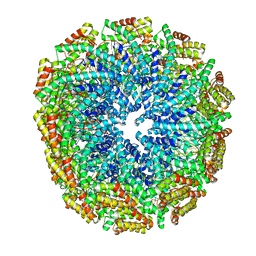 | | An envelope of a filamentous hyperthermophilic virus carries lipids in a horseshoe conformation | | 分子名称: | DNA (253-MER), ORF132, ORF140 | | 著者 | Kasson, P, DiMaio, F, Yu, X, Lucas-Staat, S, Krupovic, M, Schouten, S, Prangishvili, D, Egelman, E. | | 登録日 | 2017-06-19 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Model for a novel membrane envelope in a filamentous hyperthermophilic virus.
Elife, 6, 2017
|
|
7RMK
 
 | | Yeast CTP Synthase (Ura7) Bundle bound to substrates at low pH | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, URIDINE 5'-TRIPHOSPHATE | | 著者 | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | 登録日 | 2021-07-27 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RMF
 
 | | Substrate-bound Ura7 filament at low pH | | 分子名称: | CTP synthase | | 著者 | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | 登録日 | 2021-07-27 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (7.3 Å) | | 主引用文献 | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RMV
 
 | | Yeast CTP Synthase (Ura7) H360R Filament bound to Substrates | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, URIDINE 5'-TRIPHOSPHATE | | 著者 | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | 登録日 | 2021-07-28 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RNL
 
 | | Yeast CTP Synthase (Ura7) H360R Filament bound to Substrates | | 分子名称: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | 著者 | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | 登録日 | 2021-07-29 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
6OG1
 
 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, pre-state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Hyperactive disaggregase ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | 登録日 | 2019-04-01 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
