4LT0
 
 | | HEWL co-crystallized with Carboplatin in non-NaCl conditions: crystal 1 processed using the EVAL software package | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carboplatin binding to histidine.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4YSR
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 16.6 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
1QZ1
 
 | | Crystal Structure of the Ig 1-2-3 fragment of NCAM | | Descriptor: | Neural cell adhesion molecule 1, 140 kDa isoform | | Authors: | Soroka, V, Kolkova, K, Kastrup, J.S, Diederichs, K, Breed, J, Kiselyov, V.V, Poulsen, F.M, Larsen, I.K, Welte, W, Berezin, V, Bock, E, Kasper, C. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and interactions of NCAM Ig1-2-3 suggest a novel zipper mechanism for homophilic adhesion
Structure, 11, 2003
|
|
1YCE
 
 | | Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus | | Descriptor: | NONAN-1-OL, SODIUM ION, subunit c | | Authors: | Meier, T, Polzer, P, Diederichs, K, Welte, W, Dimroth, P. | | Deposit date: | 2004-12-22 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus.
Science, 308, 2005
|
|
1T92
 
 | | Crystal structure of N-terminal truncated pseudopilin PulG | | Descriptor: | General secretion pathway protein G, ZINC ION | | Authors: | Koehler, R, Schaefer, K, Mueller, S, Vignon, G, Diederichs, K, Philippsen, A, Ringler, P, Pugsley, A.P, Engel, A, Welte, W. | | Deposit date: | 2004-05-14 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and assembly of the pseudopilin PulG.
Mol.Microbiol., 54, 2004
|
|
4AML
 
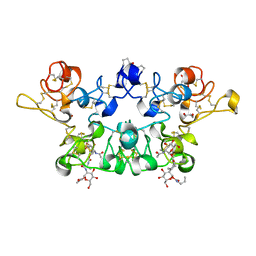 | | CRYSTAL STRUCTURE OF WHEAT GERM AGGLUTININ ISOLECTIN 1 IN COMPLEX WITH GLYCOSYLURETHAN | | Descriptor: | 2-acetamido-2-deoxy-1-O-(propylcarbamoyl)-alpha-D-glucopyranose, AGGLUTININ ISOLECTIN 1, GLYCEROL | | Authors: | Schwefel, D, Maierhofer, C, Beck, J.G, Seeberger, S, Diederichs, K, Moeller, H.M, Welte, W, Wittmann, V. | | Deposit date: | 2012-03-12 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
1URS
 
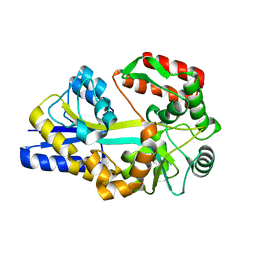 | | X-ray structures of the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-11-04 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins
J.Mol.Biol., 335, 2004
|
|
1URD
 
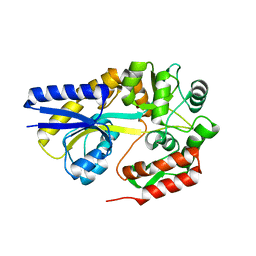 | | X-ray structures of the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius provide insight into acid stability of proteins | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-10-29 | | Release date: | 2003-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins.
J.Mol.Biol., 335, 2004
|
|
1URG
 
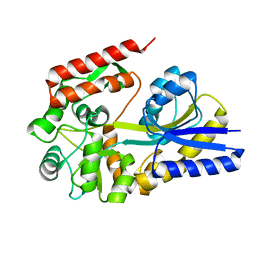 | | X-ray structures from the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-10-29 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins.
J.Mol.Biol., 335, 2004
|
|
3CSN
 
 | |
3CSL
 
 | |
4X5U
 
 | | X-ray crystal structure of CagL at pH 4.2 | | Descriptor: | Cag pathogenicity island protein (Cag18) | | Authors: | Sundberg, E.J, Bonsor, D.A, Diederichs, K. | | Deposit date: | 2014-12-05 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Integrin Engagement by the Helical RGD Motif of the Helicobacter pylori CagL Protein Is Regulated by pH-induced Displacement of a Neighboring Helix.
J.Biol.Chem., 290, 2015
|
|
3DDR
 
 | |
3DZM
 
 | |
4XJB
 
 | | X-ray structure of Lysozyme1 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJG
 
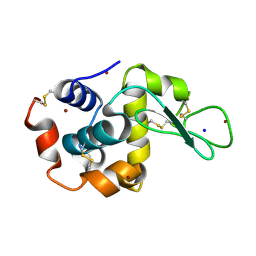 | | X-ray structure of Lysozyme B2 | | Descriptor: | BROMIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJI
 
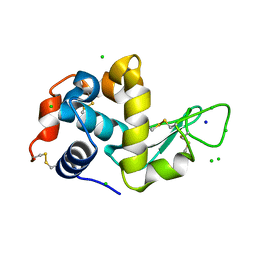 | | X-ray structure of LysozymeS2 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJD
 
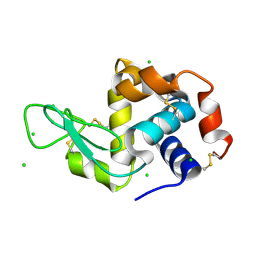 | | X-ray structure of Lysozyme2 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJH
 
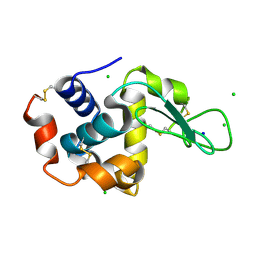 | | X-ray structure of LysozymeS1 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XNJ
 
 | | X-ray structure of PepTst2 | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Di-or tripeptide:H+ symporter, PHOSPHATE ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJF
 
 | | X-ray structure of Lysozyme B1 | | Descriptor: | BROMIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XNL
 
 | | X-ray structure of AlgE2 | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Ma, P, Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XXH
 
 | | TREHALOSE REPRESSOR FROM ESCHERICHIA COLI | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, HTH-type transcriptional regulator TreR | | Authors: | Hars, U, Horlacher, R, Boos, W, Smart, O.S, Bricogne, G, Welte, W, Diederichs, K. | | Deposit date: | 2015-01-30 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CRYSTAL STRUCTURE OF THE EFFECTOR-BINDING DOMAIN OF THE TREHALOSE-REPRESSOR OF ESCHERICHIA COLI, A MEMBER OF THE LACI FAMILY, IN ITS COMPLEXES WITH INDUCER TREHALOSE-6-PHOSPHATE AND NONINDUCER TREHALOSE.
PROTEIN SCI., 7, 1998
|
|
4XNI
 
 | | X-ray structure of PepTst1 | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Di-or tripeptide:H+ symporter, PHOSPHATE ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
