3H4I
 
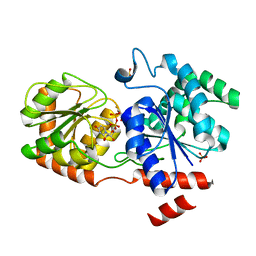 | | Chimeric Glycosyltransferase for the generation of novel natural products | | Descriptor: | Glycosyltransferase GtfA, Glycosyltransferase, SULFATE ION, ... | | Authors: | Dias, M.V.B, Truman, A.W, Wu, S, Blundell, T.L, Huang, F, Spencer, J.B. | | Deposit date: | 2009-04-20 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Chimeric glycosyltransferases for the generation of hybrid glycopeptides
Chem.Biol., 16, 2009
|
|
3KVU
 
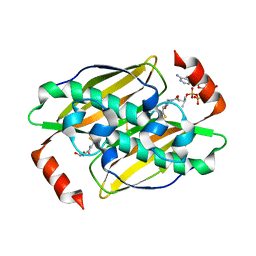 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA FlK - T42S mutant in complex with Acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Fluoroacetyl-CoA thioesterase FlK | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KVZ
 
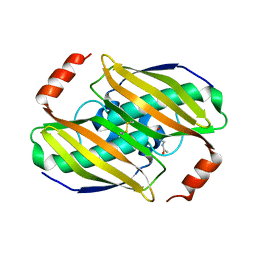 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thiesterase FlK - wild type FlK in complex with FAcCPan | | Descriptor: | (2R)-N-{3-[(5-fluoro-4-oxopentyl)amino]-3-oxopropyl}-2,4-dihydroxy-3,3-dimethylbutanamide, Fluoroacetyl-CoA thioesterase FlK | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KVI
 
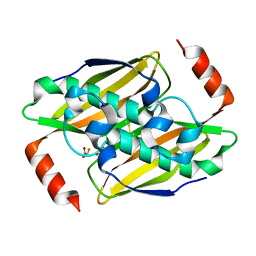 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK - T42A mutant in complex with fluoro-acetate | | Descriptor: | Fluoroacetyl-CoA thioesterase FlK, fluoroacetic acid | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KW1
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA FlK - Wild type FlK in complex with FAcOPan | | Descriptor: | 2-({N-[(2S)-2,4-dihydroxy-3,3-dimethylbutanoyl]-beta-alanyl}amino)ethyl fluoroacetate, Fluoroacetyl-CoA thioesterase | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KV8
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK - Wild type FlK in complex with fluoro-acetate | | Descriptor: | Fluoroacetyl-CoA thioesterase FlK, fluoroacetic acid | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-29 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KX7
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA FlK - apo wild type FlK | | Descriptor: | Fluoroacetyl-CoA thioesterase FlK | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-12-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KUW
 
 | | Structural basis of the activity ans substrate specificity of the fluoroacetyl-CoA thioesterase FlK - T42S mutant in complex with Fluoro-acetate | | Descriptor: | Fluoroacetyl coenzyme A thioesterase, fluoroacetic acid | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KV7
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK - wild type FlK in complex with acetate | | Descriptor: | ACETATE ION, fluoroacetyl-CoA thioesterase FlK | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-29 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KUV
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK - T42S mutant in complex with acetate. | | Descriptor: | ACETATE ION, Fluoroacetyl coenzyme A thioesterase | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
6N5H
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 5 | | Descriptor: | 4-[(trans-4-{[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylcarbamoyl]amino}cyclohexyl)oxy]benzoic acid, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriano, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors
Int. J. Biol. Macromol., 129, 2019
|
|
1WE2
 
 | | Crystal structure of shikimate kinase from mycobacterium tuberculosis in complex with MGADP and shikimic acid | | Descriptor: | 3-DEHYDROSHIKIMATE, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Pereira, J.H, de Oliveira, J.S, Canduri, F, Dias, M.V, Palma, M.S, Basso, L.A, Santos, D.S, de Azevedo Jr, W.F. | | Deposit date: | 2004-05-22 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of shikimate kinase from Mycobacterium tuberculosis reveals the binding of shikimic acid.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1V2H
 
 | | Crystal structure of human PNP complexed with guanine | | Descriptor: | GUANINE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | De Azevedo Jr, W.F, Canduri, F, Pereira, J.H, Dos Santos, D.M, Bertacine Dias, M.V, Silva, R.G, Palma, M.S, Basso, L.A, Santos, D.S. | | Deposit date: | 2003-10-16 | | Release date: | 2004-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human PNP complexed with guanine.
Biochem.Biophys.Res.Commun., 312, 2003
|
|
6N3Z
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 4 | | Descriptor: | Epoxide hydrolase TrEH, N-methyl-4-{[trans-4-({[4-(trifluoromethoxy)phenyl]carbamoyl}amino)cyclohexyl]oxy}benzamide | | Authors: | de Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6N5F
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 3 | | Descriptor: | Epoxide hydrolase TrEH, N-(8-amino-8-oxooctyl)nonanamide | | Authors: | Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6N5G
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 2 | | Descriptor: | 4-[(quinolin-3-yl)methyl]-N-[4-(trifluoromethoxy)phenyl]piperidine-1-carboxamide, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6N3K
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 1 | | Descriptor: | Epoxide hydrolase, N-{cis-4-[(2,6-difluorophenyl)methoxy]cyclohexyl}-N'-(3-phenylpropyl)urea | | Authors: | de Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6D5A
 
 | |
6WUQ
 
 | | Crystal structure of AjiA1 in apo form | | Descriptor: | AjiA1, MAGNESIUM ION, ZINC ION | | Authors: | Paiva, F.C.R, Chan, K, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2020-05-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The crystal structure of AjiA1 reveals a novel structural motion mechanism in the adenylate-forming enzyme family
Acta Crystallogr.,Sect.D, 76, 2020
|
|
8G6P
 
 | | Crystal structure of Mycobacterium thermoresistibile MurE in complex with ADP and 2,6-Diaminopimelic acid | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Rossini, N.O, Silva, C.S, Dias, M.V.B. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of Mycobacterium thermoresistibile MurE ligase reveals the binding mode of the substrate m-diaminopimelate.
J.Struct.Biol., 215, 2023
|
|
3BAF
 
 | | Crystal structure of shikimate kinase from Mycobacterium tuberculosis in complex with AMP-PNP | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Shikimate kinase | | Authors: | Faim, L.M, Dias, M.V.B, Vasconcelos, I.G, Basso, L.A, Santos, D.S, Azevedo, W.F, Ruggiero, N.J. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure for shikimate kinase from Mycobacterium tuberculosis in complex with AMP-PNP
To be Published
|
|
6D4K
 
 | |
6D51
 
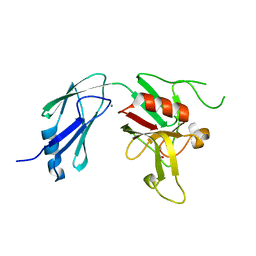 | | Crystal structure of L,D-transpeptidase 3 from Mycobacterium tuberculosis in complex with a faropenem-derived adduct | | Descriptor: | ACETYL GROUP, CALCIUM ION, Probable L,D-transpeptidase 3 | | Authors: | Libreros, G.A, Dias, M.V.B. | | Deposit date: | 2018-04-19 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for the Interaction and Processing of beta-Lactam Antibiotics by l,d-Transpeptidase 3 (LdtMt3) from Mycobacterium tuberculosis.
ACS Infect Dis, 5, 2019
|
|
3N8K
 
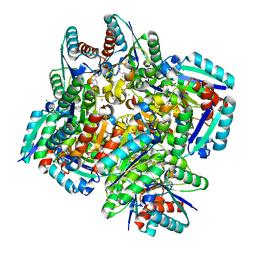 | | Type II dehydroquinase from Mycobacterium tuberculosis complexed with citrazinic acid | | Descriptor: | 2,6-dioxo-1,2,3,6-tetrahydropyridine-4-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Snee, W.C, Palaninathan, S.K, Sacchettini, J.C, Dias, M.V.B, Bromfield, K.M, Payne, R, Ciulli, A, Howard, N.I, Abell, C, Blundell, T.L, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
9AU3
 
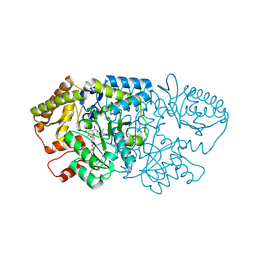 | | Crystal structure of GenB2 in complex with G418 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 6'-epimerase, C-6' aminotransferase, ... | | Authors: | De Oliveira, G.S, Bury, P.S, Huang, F, Li, Y, Araujo, N.C, Zhou, J, Sun, Y, Leeper, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2024-02-28 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Basis of GenB2 Isomerase Activity from Gentamicin Biosynthesis.
Acs Chem.Biol., 2024
|
|
