5L1D
 
 | |
3ZV4
 
 | | CRYSTAL STRUCTURE OF CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE (BPHB) FROM PANDORAEA PNOMENUSA STRAIN B-356 IN APO FORM AT 1.8 ANGSTROM | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
3ZV3
 
 | | CRYSTAL STRUCTURE OF CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE (BPHB)FROM PANDORAEA PNOMENUSA STRAIN B-356 IN INTERMEDIATE STATE OF SUBSTRATE BINDING LOOP | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
3ZV5
 
 | | CRYSTAL STRUCTURE OF CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE (BPHB) FROM PANDORAEA PNOMENUSA STRAIN B-356 COMPLEX WITH CO-ENZYME NAD AND PRODUCT 2,3-DIHYDROXYBIPHENYL | | Descriptor: | BIPHENYL-2,3-DIOL, CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
3ZV6
 
 | | CRYSTAL STRUCTURE OF CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE (BPHB) FROM PANDORAEA PNOMENUSA STRAIN B-356 COMPLEX WITH CO-ENZYME NAD AND PRODUCT ANALOG 4,4'-DIHYDROXYBIPHENYL | | Descriptor: | 4,4'-DIHYDROXYBIPHENYL, CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
5AEU
 
 | | Crystal structure of II9 variant of Biphenyl dioxygenase from Burkholderia xenovorans LB400 | | Descriptor: | BIPHENYL DIOXYGENASE SUBUNIT ALPHA, BIPHENYL DIOXYGENASE SUBUNIT BETA, FE (II) ION, ... | | Authors: | Dhindwal, S, Gomez-Gil, L, Sylvestre, M, Eltis, L.D, Bolin, J.T, Kumar, P. | | Deposit date: | 2015-01-10 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Basis of the Enhanced Pollutant-Degrading Capabilities of an Engineered Biphenyl Dioxygenase
J.Bacteriol., 198, 2016
|
|
5AEW
 
 | | Crystal structure of II9 variant of Biphenyl dioxygenase from Burkholderia xenovorans LB400 in complex with biphenyl | | Descriptor: | BIPHENYL, BIPHENYL DIOXYGENASE SUBUNIT ALPHA, BIPHENYL DIOXYGENASE SUBUNIT BETA, ... | | Authors: | Dhindwal, S, Gomez-Gil, L, Sylvestre, M, Eltis, L.D, Bolin, J.T, Kumar, P. | | Deposit date: | 2015-01-10 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis of the Enhanced Pollutant-Degrading Capabilities of an Engineered Biphenyl Dioxygenase.
J.Bacteriol., 198, 2016
|
|
2Y99
 
 | | Crystal Structure of cis-Biphenyl-2,3-dihydrodiol-2,3-dehydrogenase (BphB)from Pandoraea pnomenusa strain B-356 complex with co-enzyme NAD | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-02-12 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
2Y93
 
 | | Crystal Structure of cis-Biphenyl-2,3-dihydrodiol-2,3-dehydrogenase (BphB)from Pandoraea pnomenusa strain B-356. | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-02-11 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
4UME
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion and KDO molecule | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, MAGNESIUM ION | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-16 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UMD
 
 | |
4UM7
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase (kdsC) from Moraxella catarrhalis in complex with Magnesium ion | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, MAGNESIUM ION | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UM5
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion and Phosphate ion | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, MAGNESIUM ION, ... | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UMF
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion, Phosphate ion and KDO molecule | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, MAGNESIUM ION, ... | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-16 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5WXU
 
 | | 11S globulin from Wrightia tinctoria reveals auxin binding site | | Descriptor: | 11S globulin, 1H-INDOL-3-YLACETIC ACID, CITRATE ANION, ... | | Authors: | Kumar, P, Kesari, P, Dhindwal, S, Kumar, P. | | Deposit date: | 2017-01-09 | | Release date: | 2018-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel function for globulin in sequestering plant hormone: Crystal structure of Wrightia tinctoria 11S globulin in complex with auxin.
Sci Rep, 7, 2017
|
|
6DZU
 
 | |
5FTW
 
 | | Crystal structure of glutamate O-methyltransferase in complex with S- adenosyl-L-homocysteine (SAH) from Bacillus subtilis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHEMOTAXIS PROTEIN METHYLTRANSFERASE, GLYCEROL, ... | | Authors: | Sharma, R, Dhindwal, S, Batra, M, Aggarwal, M, Kumar, P, Tomar, S. | | Deposit date: | 2016-01-18 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Pentapeptide-Independent Chemotaxis Receptor Methyltransferase (Cher) Reveals Idiosyncratic Structural Determinants for Receptor Recognition.
J.Struct.Biol., 196, 2016
|
|
3ZHW
 
 | | X-ray Crystallographic Structural Characteristics of Arabidopsis Hemoglobin I and their Functional Implications | | Descriptor: | NON-SYMBIOTIC HEMOGLOBIN 1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Mukhi, N, Dhindwal, S, Uppal, S, Kumar, P, Kaur, J, Kundu, S. | | Deposit date: | 2012-12-30 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | X-Ray Crystallographic Structural Characteristics of Arabidopsis Hemoglobin I and Their Functional Implications
Biochim.Biophys.Acta, 1834, 2013
|
|
6E2R
 
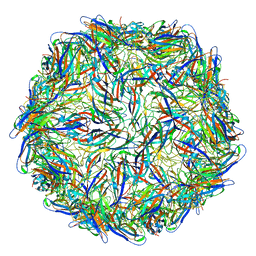 | | Mechanism of cellular recognition by PCV2 | | Descriptor: | Capsid protein of PCV2 | | Authors: | Khayat, R, Dhindwal, S. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Porcine Circovirus 2 Uses a Multitude of Weak Binding Sites To Interact with Heparan Sulfate, and the Interactions Do Not Follow the Symmetry of the Capsid.
J.Virol., 93, 2019
|
|
6E32
 
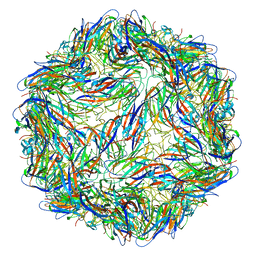 | | Capsid protein of PCV2 with N,O6-DISULFO-GLUCOSAMINE | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Capsid protein of PCV2 | | Authors: | Khayat, R, Dhindwal, S. | | Deposit date: | 2018-07-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Porcine Circovirus 2 Uses a Multitude of Weak Binding Sites To Interact with Heparan Sulfate, and the Interactions Do Not Follow the Symmetry of the Capsid.
J.Virol., 93, 2019
|
|
6E34
 
 | | Capsid protein of PCV2 with N,O6-DISULFO-GLUCOSAMINE and 2-O-sulfo-alpha-L-idopyranuronic acid | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Capsid protein of PCV2 | | Authors: | Khayat, R, Dhindwal, S. | | Deposit date: | 2018-07-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Porcine Circovirus 2 Uses a Multitude of Weak Binding Sites To Interact with Heparan Sulfate, and the Interactions Do Not Follow the Symmetry of the Capsid.
J.Virol., 93, 2019
|
|
6E39
 
 | |
6E30
 
 | | Mechanism of cellular recognition by PCV2 | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Capsid protein of PCV2 | | Authors: | Khayat, R, Dhindwal, S. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Porcine Circovirus 2 Uses a Multitude of Weak Binding Sites To Interact with Heparan Sulfate, and the Interactions Do Not Follow the Symmetry of the Capsid.
J.Virol., 93, 2019
|
|
6E2X
 
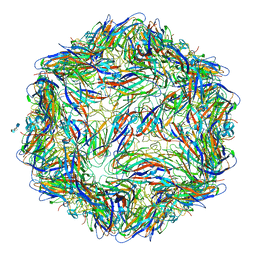 | | Mechanism of cellular recognition by PCV2 | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Capsid protein of PCV2 | | Authors: | Khayat, R, Dhindwal, S. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Porcine Circovirus 2 Uses a Multitude of Weak Binding Sites To Interact with Heparan Sulfate, and the Interactions Do Not Follow the Symmetry of the Capsid.
J.Virol., 93, 2019
|
|
6E2Z
 
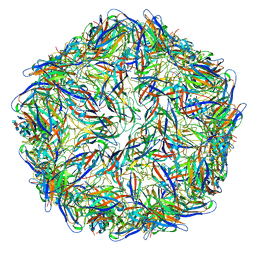 | | Mechanism of cellular recognition by PCV2 | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Capsid protein of PCV2 | | Authors: | Khayat, R, Dhindwal, S. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Porcine Circovirus 2 Uses a Multitude of Weak Binding Sites To Interact with Heparan Sulfate, and the Interactions Do Not Follow the Symmetry of the Capsid.
J.Virol., 93, 2019
|
|
