1SQZ
 
 | | Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between Group II Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution | | Descriptor: | Phospholipase A2, SULFATE ION, synthetic peptide | | Authors: | Singh, N, Prem Kumar, R, Somvanshi, R.K, Bilgrami, S, Ethayathulla, A.S, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2004-03-22 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between GroupII Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution
To be Published
|
|
1TDV
 
 | | Non-specific binding to phospholipase A2:Crystal structure of the complex of PLA2 with a designed peptide Tyr-Trp-Ala-Ala-Ala-Ala at 1.7A resolution | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa, SULFATE ION, YWAAAA | | Authors: | Singh, N, Jabeen, T, Ethayathulla, A.S, Somvanshi, R.K, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-05-24 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Non-specific binding to phospholipase A2:Crystal structure of the complex of PLA2 with a designed peptide Tyr-Trp-Ala-Ala-Ala-Ala at 1.7A resolution
to be published
|
|
1T37
 
 | | Design of specific inhibitors of phospholipase A2: Crystal structure of the complex formed between group I phospholipase A2 and a designed pentapeptide Leu-Ala-Ile-Tyr-Ser at 2.6A resolution | | Descriptor: | ACETATE ION, Phospholipase A2 isoform 3, Synthetic peptide | | Authors: | Singh, R.K, Singh, N, Jabeen, T, Makker, J, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2004-04-25 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of group I PLA2 with a group II-specific peptide Leu-Ala-Ile-Tyr-Ser (LAIYS) at 2.6 A resolution.
J.Drug Target., 13, 2005
|
|
1SKG
 
 | | Structure-based rational drug design: Crystal structure of the complex formed between Phospholipase A2 and a pentapeptide Val-Ala-Phe-Arg-Ser | | Descriptor: | METHANOL, Phospholipase A2, SULFATE ION, ... | | Authors: | Ethayathulla, A.S, Singh, N, Sharma, S, Makker, J, Dey, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure-based rational drug design: Crystal structure of the complex formed between Phospholipase A2 and a pentapeptide Val-Ala-Phe-Arg-Ser
To be Published
|
|
1TG1
 
 | | Crystal Structure of the complex formed between russells viper phospholipase A2 and a designed peptide inhibitor PHQ-Leu-Val-Arg-Tyr at 1.2A resolution | | Descriptor: | ACETIC ACID, METHANOL, Phospholipase A2, ... | | Authors: | Singh, N, Kaur, P, Somvanshi, R.K, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-05-28 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of the complex formed between russells viper phospholipase A2 and a designed peptide inhibitor Cbz-dehydro-Leu-Val-Arg-Tyr at 1.2A resolution
To be Published
|
|
1TJK
 
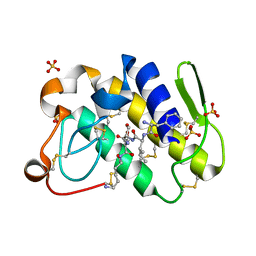 | | Crystal structure of the complex formed between group II phospholipase A2 with a designed pentapeptide, Phe- Leu- Ser- Thr- Lys at 1.2 A resolution | | Descriptor: | Phospholipase A2, SULFATE ION, synthetic peptide | | Authors: | Singh, N, Jabeen, T, Somvanshi, R.K, Sharma, S, Perbandt, M, Dey, S, Betzel, C, Singh, T.P. | | Deposit date: | 2004-06-06 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the complex formed between group II phospholipase A2 with a designed pentapeptide, Phe - Leu - Ser - Thr - Lys at 1.2 A resolution
To be Published
|
|
1TJ9
 
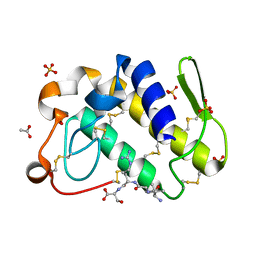 | | Structure of the complexed formed between group II phospholipase A2 and a rationally designed tetra peptide,Val-Ala-Arg-Ser at 1.1A resolution | | Descriptor: | ACETIC ACID, Phospholipase A2, SULFATE ION, ... | | Authors: | Singh, N, Ethayathulla, A.S, K Somvanshi, R, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Kaur, P, Singh, T.P. | | Deposit date: | 2004-06-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of the complex formed between group II phospholipase A2 and a rationally designed tetra peptide,Val-Ala-Arg-Ser at 1.1A resolution
TO BE PUBLISHED
|
|
1TG4
 
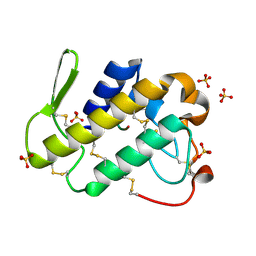 | | Design of specific inhibitors of groupII phospholipase A2(PLA2): Crystal structure of the complex formed between russells viper PLA2 and designed peptide Phe-Leu-Ala-Tyr-Lys at 1.7A resolution | | Descriptor: | FLAYK peptide, Phospholipase A2, SULFATE ION | | Authors: | Singh, N, Somvanshi, R.K, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Ethayathulla, A.S, Singh, T.P. | | Deposit date: | 2004-05-28 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of specific inhibitors of groupII phospholipase A2(PLA2): Crystal structure of the complex formed between russells viper PLA2 and designed peptide Phe-Leu-Ala-Tyr-Lys at 1.7A resolution
TO BE PUBLISHED
|
|
4LT4
 
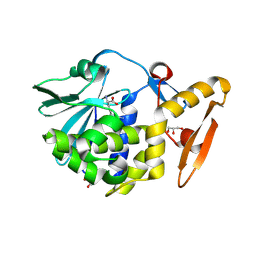 | | Crystal structure of arginine inhibited Ribosome inactivating protein from Momordica balsamina at 1.69 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, GLYCEROL, ... | | Authors: | Yamini, S, Pandey, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of arginine inhibited Ribosome inactivating protein from Momordica balsamina at 1.69 A resolution
To be Published
|
|
5WV1
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with ribose sugar at 1.90 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Shokeen, A, Singh, P.K, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with ribose sugar at
1.90 A resolution.
To Be Published
|
|
7AZT
 
 | | X-ray crystallographic structure of (6-4)photolyase from Drosophila melanogaster at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, RE11660p | | Authors: | Cellini, A, Wahlgren, W.Y, Henry, L, Westenhoff, S, Pandey, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6T3L
 
 | | PAS-GAF fragment from Deinococcus radiodurans phytochrome in dark state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Yuan Wahlgren, W, Pandey, S, Schmidt, M, Westenhoff, S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser.
Elife, 9, 2020
|
|
6T3U
 
 | | PAS-GAF fragment from Deinococcus radiodurans phytochrome 1ps after photoexcitation | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Yuan Wahlgren, W, Pandey, S, Schmidt, M, Westenhoff, S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser.
Elife, 9, 2020
|
|
5CSO
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleoside, cytidine at 1.78 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
5CST
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleotide, cytidine diphosphate at 1.78 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
4O4Q
 
 | | Crystal structure of the complex formed between type 1 ribosome inactivating protein and uridine diphosphate at 1.81 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Yamini, S, Pandey, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the complex formed between type 1 ribosome inactivating protein and uridine diphosphate at 1.81 A resolution
To be Published
|
|
4O0O
 
 | | Crystal structure of the complex of type 1 Ribosome inactivating protein from Momordica balsamina with 5-fluorouracil at 2.59 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-FLUOROURACIL, GLYCEROL, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-12-14 | | Release date: | 2014-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of the complex of type 1 Ribosome inactivating protein from Momordica balsamina with 5-fluorouracil at 2.59 A resolution
To be Published
|
|
5CIX
 
 | | Structure of the complex of type 1 Ribosome inactivating protein with triethanolamine at 1.88 Angstrom resolution | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Singh, P.K, Pandey, S, Tyagi, T.K, Singh, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-13 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure of the complex of type 1 Ribosome inactivating protein with triethanolamine at 1.88 Angstrom resolution.
To Be Published
|
|
4KPV
 
 | | Crystal structure of the complex of ribosome inactivating protein from Momordica balsamina with Pyrimidine-2,4(1H,3H)-dione at 2.57 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, URACIL, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-14 | | Release date: | 2013-05-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of the complex of ribosome inactivating protein from Momordica balsamina with Pyrimidine-2,4(1H,3H)-dione at 2.57 A resolution
To be Published
|
|
4K2Z
 
 | | Crystal structure of the complex of type I Ribosome inactivating protein from Momordica balsamina with Methylethylamine at 1.80 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, METHYLETHYLAMINE, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex of type I Ribosome inactivating protein from Momordica balsamina with Methylethylamine at 1.80 A resolution
To be Published
|
|
4KWN
 
 | | A new stabilizing water structure at the substrate binding site in ribosome inactivating protein from Momordica balsamina at 1.80 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Singh, A, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-24 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A new stabilizing water structure at the substrate binding site in ribosome inactivating protein from Momordica balsamina at 1.80 A resolution
To be Published
|
|
4KMK
 
 | | Crystal structure of Ribosome Inactivating protein from Momordica balsamina at 1.65 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Ribosome Inactivating protein from Momordica balsamina at 1.65 A resolution
To be Published
|
|
4L66
 
 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina with highly ordered water structure in the substrate binding site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-06-12 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina with highly ordered water structure in the substrate binding site
To be Published
|
|
4LRO
 
 | | Crystal structure of spermidine inhibited Ribosome inactivating protein from Momordica balsamina | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SPERMIDINE, ... | | Authors: | Yamini, S, Pandey, S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-20 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of spermidine inhibited Ribosome inactivating protein from Momordica balsamina
To be Published
|
|
4LWX
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica Balsamina with peptidoglycan fragment at 1.78 A resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALANINE, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-29 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica Balsamina with peptidoglycan fragment at 1.78 A resolution
To be Published
|
|
