3TV0
 
 | |
1FXW
 
 | |
1CC0
 
 | | CRYSTAL STRUCTURE OF THE RHOA.GDP-RHOGDI COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, rho GDP dissociation inhibitor alpha, ... | | Authors: | Longenecker, K.L, Read, P, Derewenda, U, Dauter, Z, Garrard, S, Walker, L, Somlyo, A.V, Somlyo, A.P, Nakamoto, R.K, Derewenda, Z.S. | | Deposit date: | 1999-03-03 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | How RhoGDI binds Rho.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1MJD
 
 | | Structure of N-terminal domain of human doublecortin | | Descriptor: | DOUBLECORTIN | | Authors: | Kim, M.H, Cierpicki, T, Derewenda, U, Krowarsch, D, Feng, Y, Devedjiev, Y, Dauter, Z, Walsh, C.A, Otlewski, J, Bushweller, J.H, Derewenda, Z.S. | | Deposit date: | 2002-08-27 | | Release date: | 2003-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The DCX-domain Tandems of Doublecortin and Doublecortin-like Kinase
Nat.Struct.Biol., 10, 2003
|
|
1KMZ
 
 | | MOLECULAR BASIS OF MITOMYCIN C RESICTANCE IN STREPTOMYCES: CRYSTAL STRUCTURES OF THE MRD PROTEIN WITH AND WITHOUT A DRUG DERIVATIVE | | Descriptor: | mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
1KLL
 
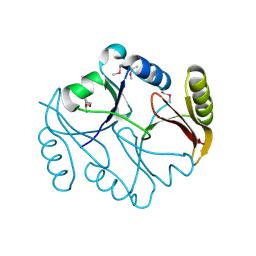 | | Molecular basis of mitomycin C resictance in streptomyces: Crystal structures of the MRD protein with and without a drug derivative | | Descriptor: | 1,2-CIS-1-HYDROXY-2,7-DIAMINO-MITOSENE, mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-12 | | Release date: | 2002-07-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
1N99
 
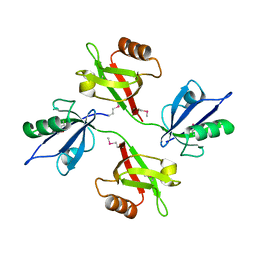 | | CRYSTAL STRUCTURE OF THE PDZ TANDEM OF HUMAN SYNTENIN | | Descriptor: | Syntenin 1 | | Authors: | Kang, B.S, Cooper, D.R, Jelen, F, Devedjiev, Y, Derewenda, U, Dauter, Z, Otlewski, J, Derewenda, Z.S. | | Deposit date: | 2002-11-22 | | Release date: | 2003-04-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | PDZ Tandem of Human Syntenin: Crystal Structure and Functional Properties
Structure, 11, 2003
|
|
4TGL
 
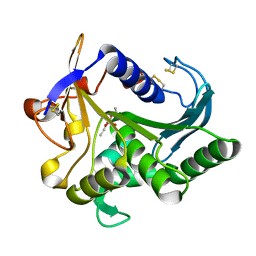 | | CATALYSIS AT THE INTERFACE: THE ANATOMY OF A CONFORMATIONAL CHANGE IN A TRIGLYCERIDE LIPASE | | Descriptor: | DIETHYL PHOSPHONATE, TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Derewenda, U, Brzozowski, A.M, Lawson, D, Derewenda, Z.S. | | Deposit date: | 1991-07-29 | | Release date: | 1993-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis at the interface: the anatomy of a conformational change in a triglyceride lipase.
Biochemistry, 31, 1992
|
|
1XCG
 
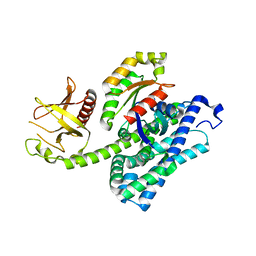 | | Crystal Structure of Human RhoA in complex with DH/PH fragment of PDZRHOGEF | | Descriptor: | Rho guanine nucleotide exchange factor 11, Transforming protein RhoA | | Authors: | Derewenda, U, Oleksy, A, Stevenson, A.S, Korczynska, J, Dauter, Z, Somlyo, A.P, Otlewski, J, Somlyo, A.V, Derewenda, Z.S. | | Deposit date: | 2004-09-01 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of RhoA in complex with the DH/PH fragment of PDZRhoGEF, an activator of the Ca(2+) sensitization pathway in smooth muscle
Structure, 12, 2004
|
|
1VZY
 
 | | Crystal structure of the Bacillus subtilis HSP33 | | Descriptor: | 33 KDA CHAPERONIN, ACETATE ION, ZINC ION | | Authors: | Janda, I.K, Devedjiev, Y, Derewenda, U, Dauter, Z, Bielnicki, J, Cooper, D.R, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-29 | | Release date: | 2004-10-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of the reduced, Zn2+-bound form of the B. subtilis Hsp33 chaperone and its implications for the activation mechanism.
Structure, 12, 2004
|
|
1WAB
 
 | | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | ACETATE ION, PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Swenson, L, Derewenda, U, Serre, L, Wei, Y, Dauter, Z, Hattori, M, Aoki, J, Arai, H, Adachi, T, Inoue, K, Derewenda, Z.S. | | Deposit date: | 1996-10-30 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Brain acetylhydrolase that inactivates platelet-activating factor is a G-protein-like trimer.
Nature, 385, 1997
|
|
1E0W
 
 | | Xylanase 10A from Sreptomyces lividans. native structure at 1.2 angstrom resolution | | Descriptor: | ENDO-1,4-BETA-XYLANASE A | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1Y7M
 
 | | Crystal Structure of the B. subtilis YkuD protein at 2 A resolution | | Descriptor: | CADMIUM ION, SULFATE ION, hypothetical protein BSU14040 | | Authors: | Bielnicki, J.A, Devedjiev, Y, Derewenda, U, Dauter, Z, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-09 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | B. subtilis ykuD protein at 2.0 A resolution: insights into the structure and function of a novel, ubiquitous family of bacterial enzymes.
Proteins, 62, 2006
|
|
1E0V
 
 | | Xylanase 10A from Sreptomyces lividans. cellobiosyl-enzyme intermediate at 1.7 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1E0X
 
 | | XYLANASE 10A FROM SREPTOMYCES LIVIDANS. XYLOBIOSYL-ENZYME INTERMEDIATE AT 1.65 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
6B6Z
 
 | | Crystal structure of the Apo Antibody fragment (Fab) raised against C-terminal domain of Ebola nucleoprotein (EBOV, TAFV, BDBV strains) | | Descriptor: | Apo Fab Heavy Chain, Apo Fab Light Chain, ZINC ION | | Authors: | Radwanska, M.J, Derewenda, U, Kossiakoff, A.A, Derewenda, Z.S. | | Deposit date: | 2017-10-03 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Crystal structure of the Apo Antibody fragment (Fab) raised against C-terminal domain of Ebola nucleoprotein (EBOV, TAFV, BDBV strains)
To Be Published
|
|
6INS
 
 | | X-RAY ANALYSIS OF THE SINGLE CHAIN B29-A1 PEPTIDE-LINKED INSULIN MOLECULE. A COMPLETELY INACTIVE ANALOGUE | | Descriptor: | INSULIN, ZINC ION | | Authors: | Derewenda, U, Derewenda, Z, Dodson, E.J, Dodson, G.G, Bing, X, Markussen, J. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analysis of the single chain B29-A1 peptide-linked insulin molecule. A completely inactive analogue.
J.Mol.Biol., 220, 1991
|
|
5DSD
 
 | | The crystal structure of the C-terminal domain of Ebola (Bundibugyo) nucleoprotein | | Descriptor: | CHLORIDE ION, GLYCEROL, Nucleoprotein | | Authors: | Baker, L, Handing, K.B, Utepbergenov, D, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2015-09-17 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Molecular architecture of the nucleoprotein C-terminal domain from the Ebola and Marburg viruses.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5E2X
 
 | | The crystal structure of the C-terminal domain of Ebola (Tai Forest) nucleoprotein | | Descriptor: | NONAETHYLENE GLYCOL, NP | | Authors: | Baker, L.E, Handing, K.B, Derewenda, U, Utepbergenov, D, Derewenda, Z.S. | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular architecture of the nucleoprotein C-terminal domain from the Ebola and Marburg viruses.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2V71
 
 | | Coiled-coil region of NudEL | | Descriptor: | NUCLEAR DISTRIBUTION PROTEIN NUDE-LIKE 1 | | Authors: | Derewenda, U, Cooper, D.R, Kim, M.H, Derewenda, Z.S. | | Deposit date: | 2007-07-25 | | Release date: | 2007-11-27 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Structure of the Coiled-Coil Domain of Ndel1 and the Basis of its Interaction with Lis1, the Causal Protein of Miller-Dieker Lissencephaly.
Structure, 15, 2007
|
|
4EL9
 
 | | Structure of N-terminal kinase domain of RSK2 with afzelin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, Ribosomal protein S6 kinase alpha-3 | | Authors: | Utepbergenov, D, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2012-04-10 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Insights into the Inhibition of the p90 Ribosomal S6 Kinase (RSK) by the Flavonol Glycoside SL0101 from the 1.5 A Crystal Structure of the N-Terminal Domain of RSK2 with Bound Inhibitor.
Biochemistry, 51, 2012
|
|
4GUE
 
 | | Structure of N-terminal kinase domain of RSK2 with flavonoid glycoside quercitrin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, MAGNESIUM ION, Ribosomal protein S6 kinase alpha-3, ... | | Authors: | Derewenda, U, Utepbergenov, D, Szukalska, G, Derewenda, Z.S. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of quercitrin as an inhibitor of the p90 S6 ribosomal kinase (RSK): structure of its complex with the N-terminal domain of RSK2 at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4QAZ
 
 | |
4QB0
 
 | |
1KMT
 
 | | Crystal structure of RhoGDI Glu(154,155)Ala mutant | | Descriptor: | Rho GDP-dissociation inhibitor 1 | | Authors: | Mateja, A, Devedjiev, Y, Krowarsh, D, Longenecker, K, Dauter, Z, Otlewski, J, Derewenda, Z.S. | | Deposit date: | 2001-12-17 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The impact of Glu-->Ala and Glu-->Asp mutations on the crystallization properties of RhoGDI: the structure of RhoGDI at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
