7YXX
 
 | |
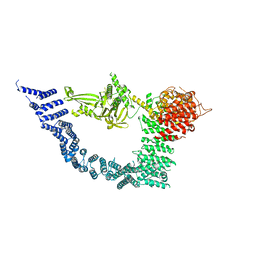
7YXY
 
 | |
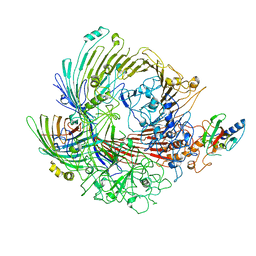
6H3J
 
 | |
6H3I
 
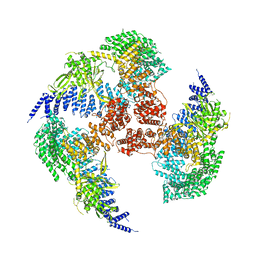
 | | Structural snapshots of the Type 9 protein translocon | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, PorV, Protein involved in gliding motility SprA | | Authors: | Deme, J.C, Lea, S.M. | | Deposit date: | 2018-07-18 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Type 9 secretion system structures reveal a new protein transport mechanism.
Nature, 564, 2018
|
|
8UCS
 
 | |
7OXP
 
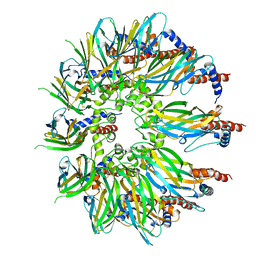 | | Cryo-EM structure of yeast Sei1 | | Descriptor: | BJ4_G0032880.mRNA.1.CDS.1,BJ4_G0032880.mRNA.1.CDS.1 | | Authors: | Deme, J.C, Lea, S.M. | | Deposit date: | 2021-06-22 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of lipid droplet formation by the yeast Sei1/Ldb16 Seipin complex.
Nat Commun, 12, 2021
|
|
7OXR
 
 | |
8GL6
 
 | |
8GLJ
 
 | |
8GLK
 
 | |
8GLN
 
 | |
8GL8
 
 | |
8GLM
 
 | |
8UPL
 
 | | Cryo-EM structure of a Clockwise locked form of the Salmonella enterica Typhimurium flagellar C-ring, with C34 symmetry applied | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2023-10-22 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
8UOX
 
 | | Cryo-EM structure of a Counterclockwise locked form of the Salmonella enterica Typhimurium flagellar C-ring, with C34 symmetry applied | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2023-10-20 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
8UMD
 
 | | Cryo-EM structure of a single subunit of a Counterclockwise-locked form of the Salmonella enterica Typhimurium flagellar C-ring. | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2023-10-17 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
8UMX
 
 | | Cryo-EM structure of a single subunit of a Clockwise-locked form of the Salmonella enterica Typhimurium flagellar C-ring. | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2023-10-18 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
7AKV
 
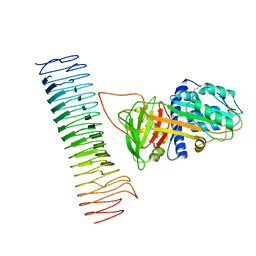 | | The cryo-EM structure of the Vag8-C1 inhibitor complex | | Descriptor: | Plasma protease C1 inhibitor, Vag8 | | Authors: | Johnson, S, Lea, S.M, Deme, J.C, Furlong, E, Dhillon, A. | | Deposit date: | 2020-10-02 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular Basis for Bordetella pertussis Interference with Complement, Coagulation, Fibrinolytic, and Contact Activation Systems: the Cryo-EM Structure of the Vag8-C1 Inhibitor Complex.
Mbio, 12, 2021
|
|
7BC6
 
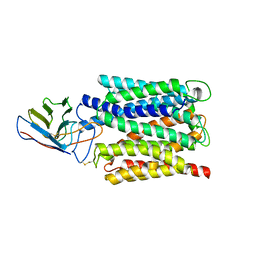 | | Cryo-EM structure of the outward open proton coupled folate transporter at pH 7.5 | | Descriptor: | Proton-coupled folate transporter, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of antifolate recognition and transport by PCFT.
Nature, 595, 2021
|
|
7BC7
 
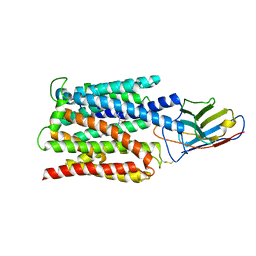 | | Cryo-EM structure of the proton coupled folate transporter at pH 6.0 bound to pemetrexed | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, Proton-coupled folate transporter, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of antifolate recognition and transport by PCFT.
Nature, 595, 2021
|
|
7P9V
 
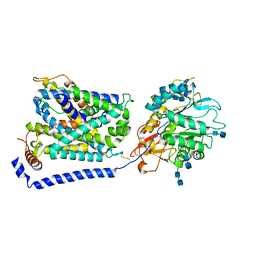 | | Cryo EM structure of System XC- | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
7P9U
 
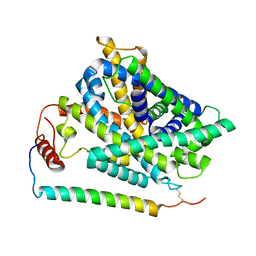 | | Cryo EM structure of System XC- in complex with glutamate | | Descriptor: | 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter, GLUTAMIC ACID | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
8FY3
 
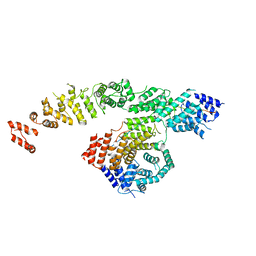 | | Structure of NOT1:NOT10:NOT11 module of the human CCR4-NOT complex | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 10, CCR4-NOT transcription complex subunit 11 | | Authors: | Lea, S.M, Deme, J.C, Raisch, T, Pekovic, F, Valkov, E. | | Deposit date: | 2023-01-25 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structure and assembly of the NOT10:11 module of the CCR4-NOT complex.
Commun Biol, 6, 2023
|
|
8FY4
 
 | | Structure of NOT1:NOT10:NOT11 module of the chicken CCR4-NOT complex | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 10, CCR4-NOT transcription complex subunit 11 | | Authors: | Lea, S.M, Deme, J.C, Raisch, T, Levdansky, Y, Valkov, E. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structure and assembly of the NOT10:11 module of the CCR4-NOT complex.
Commun Biol, 6, 2023
|
|
8SAH
 
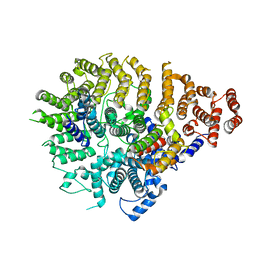 | | Huntingtin C-HEAT domain in complex with HAP40 | | Descriptor: | 40-kDa huntingtin-associated protein, Huntingtin | | Authors: | Harding, R.J, Deme, J.C, Alteen, M.G, Arrowsmith, C.H, Lea, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-31 | | Release date: | 2023-04-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Delineation of functional subdomains of Huntingtin protein and their interaction with HAP40.
Structure, 31, 2023
|
|
