7SB2
 
 | |
7SAT
 
 | |
7SAZ
 
 | |
7SAU
 
 | |
7SAX
 
 | |
7P9V
 
 | | Cryo EM structure of System XC- | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
7P9U
 
 | | Cryo EM structure of System XC- in complex with glutamate | | Descriptor: | 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter, GLUTAMIC ACID | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
6YSF
 
 | |
6YS8
 
 | |
6X9O
 
 | | High resolution cryoEM structure of huntingtin in complex with HAP40 | | Descriptor: | 40-kDa huntingtin-associated protein, Huntingtin | | Authors: | Harding, R.J, Deme, J.C, Lea, S.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Huntingtin structure is orchestrated by HAP40 and shows a polyglutamine expansion-specific interaction with exon 1.
Commun Biol, 4, 2021
|
|
6YSL
 
 | |
6R6B
 
 | | Structure of the core Shigella flexneri type III secretion system export gate complex SctRST (Spa24/Spa9/Spa29). | | Descriptor: | Surface presentation of antigens protein SpaP, Surface presentation of antigens protein SpaQ, Surface presentation of antigens protein SpaR | | Authors: | Johnson, S, Kuhlen, L, Deme, J.C, Abrusci, P, Lea, S.M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The Structure of an Injectisome Export Gate Demonstrates Conservation of Architecture in the Core Export Gate between Flagellar and Virulence Type III Secretion Systems.
Mbio, 10, 2019
|
|
8HC1
 
 | | CryoEM structure of Helicobacter pylori UreFD/urease complex | | Descriptor: | Urease accessory protein UreF, Urease accessory protein UreH, Urease subunit alpha, ... | | Authors: | Nim, Y.S, Fong, I.Y.H, Deme, J, Tsang, K.L, Caesar, J, Johnson, S, Wong, K.B, Lea, S.M. | | Deposit date: | 2022-11-01 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Delivering a toxic metal to the active site of urease.
Sci Adv, 9, 2023
|
|
8HCN
 
 | | CryoEM Structure of Klebsiella pneumoniae UreD/urease complex | | Descriptor: | Urease accessory protein UreD, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Nim, Y.S, Fong, I.Y.H, Deme, J, Tsang, K.L, Caesar, J, Johnson, S, Wong, K.B, Lea, S.M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Delivering a toxic metal to the active site of urease.
Sci Adv, 9, 2023
|
|
8SA2
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 1 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 1 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA3
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 2 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 2 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA6
 
 | | apo form of adenosylcobalamin riboswitch dimer | | Descriptor: | apo form of adenosylcobalamin riboswitch dimer | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA5
 
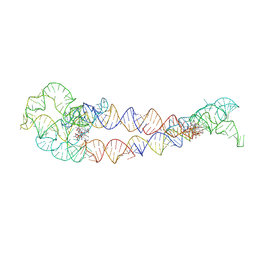 | | Adenosylcobalamin-bound riboswitch dimer, form 4 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 4 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA4
 
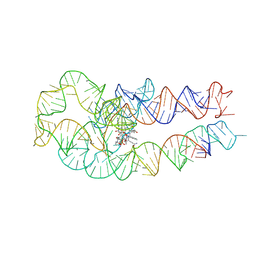 | | Adenosylcobalamin-bound riboswitch dimer, form 3 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 3 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8UPL
 
 | | Cryo-EM structure of a Clockwise locked form of the Salmonella enterica Typhimurium flagellar C-ring, with C34 symmetry applied | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2023-10-22 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
9BIT
 
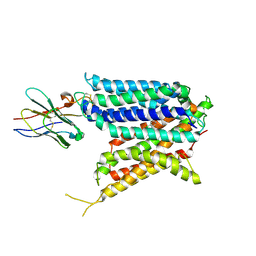 | | Cryo-EM structure of the mammalian peptide transporter PepT2 bound to cloxacillin, pose 1 | | Descriptor: | CLOXACILLIN, Solute carrier family 15 member 2, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2024-04-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for antibiotic transport and inhibition in PepT2.
Nat Commun, 15, 2024
|
|
9BIU
 
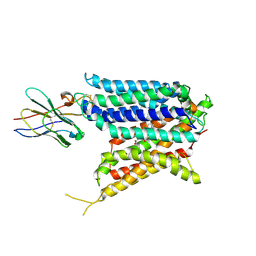 | | Cryo-EM structure of the mammalian peptide transporter PepT2 bound to cloxacillin, pose 2 | | Descriptor: | CLOXACILLIN, Solute carrier family 15 member 2, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2024-04-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for antibiotic transport and inhibition in PepT2.
Nat Commun, 15, 2024
|
|
9BIR
 
 | | Cryo-EM structure of the mammalian peptide transporter PepT2 bound to cefadroxil | | Descriptor: | Cefadroxil, Solute carrier family 15 member 2, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2024-04-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for antibiotic transport and inhibition in PepT2.
Nat Commun, 15, 2024
|
|
9BIS
 
 | | Cryo-EM structure of the mammalian peptide transporter PepT2 bound to amoxicillin | | Descriptor: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, Solute carrier family 15 member 2, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2024-04-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for antibiotic transport and inhibition in PepT2.
Nat Commun, 15, 2024
|
|
6S3L
 
 | | Structure of the core of the flagellar export apparatus from Vibrio mimicus, the FliPQR-FlhB complex. | | Descriptor: | Flagellar biosynthetic protein FlhB, Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, ... | | Authors: | Kuhlen, L, Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2019-06-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The substrate specificity switch FlhB assembles onto the export gate to regulate type three secretion.
Nat Commun, 11, 2020
|
|
