7UDY
 
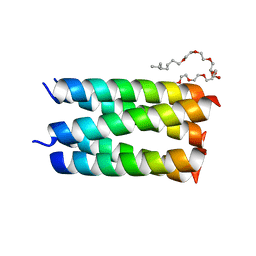 | | Designed pentameric channel QLLL | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Designed channel QLLL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
3URM
 
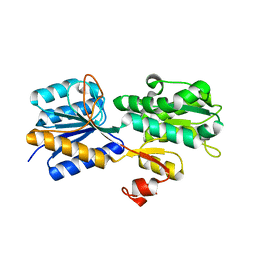 | | Crystal structure of the periplasmic sugar binding protein ChvE | | Descriptor: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-galactopyranose | | Authors: | Hu, X, Zhao, J, Binns, A, Degrado, W. | | Deposit date: | 2011-11-22 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6YB1
 
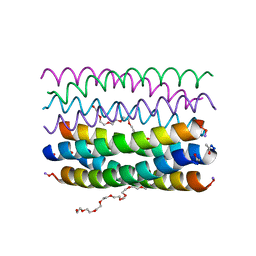 | | Crystal structure of an antiparallel octameric transmembrane coiled coil K2-CCTM-VbIc | | Descriptor: | GLYCINE, K2-CCTM-VbIc, NONAETHYLENE GLYCOL | | Authors: | Kratochvil, H.T, Liu, L, Scott, A.J, Woolfson, D.N, DeGrado, W.F. | | Deposit date: | 2020-03-15 | | Release date: | 2021-04-07 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Constructing ion channels from water-soluble alpha-helical barrels.
Nat.Chem., 13, 2021
|
|
3V86
 
 | |
4QKM
 
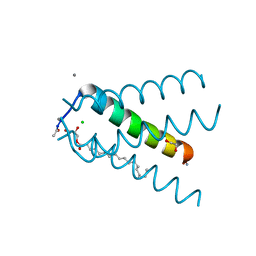 | | Influenza A M2 wild type TM domain at low pH in the lipidic cubic phase under room temperature diffraction conditions | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Thomaston, J.L, DeGrado, W.F. | | Deposit date: | 2014-06-06 | | Release date: | 2015-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | High-resolution structures of the M2 channel from influenza A virus reveal dynamic pathways for proton stabilization and transduction.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5C02
 
 | |
1YOD
 
 | |
6US9
 
 | |
6US8
 
 | |
8TNB
 
 | |
8TNC
 
 | |
8TN1
 
 | |
8TND
 
 | |
8TN6
 
 | |
6B17
 
 | | Design of a short thermally stable alpha-helix embedded in a macrocycle | | Descriptor: | 3,3'-dimethyl-1,1'-biphenyl, Capped-strapped peptide | | Authors: | Wu, H, Acharyya, A, Wu, Y, Liu, L, Jo, H, Gai, F, DeGrado, W.F. | | Deposit date: | 2017-09-17 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design of a Short Thermally Stable alpha-Helix Embedded in a Macrocycle.
Chembiochem, 19, 2018
|
|
6ANF
 
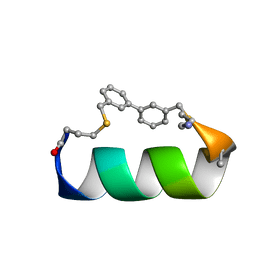 | | Design of a short thermo-stable alpha-helix embedded in a macrocycle | | Descriptor: | 3,3'-dimethyl-1,1'-biphenyl, Capped-strapped peptide | | Authors: | Wu, H, Acharyya, A, Wu, Y, Liu, L, Jo, H, Gai, F, DeGrado, W.F. | | Deposit date: | 2017-08-13 | | Release date: | 2018-02-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Design of a Short Thermally Stable alpha-Helix Embedded in a Macrocycle.
Chembiochem, 19, 2018
|
|
1MFT
 
 | | Crystal Structure Of Four-Helix Bundle Model | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Lahr, S.J, Stayrook, S.E, North, B, Kaplan, J, Geremia, S, DeGrado, W. | | Deposit date: | 2002-08-13 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
1COI
 
 | | DESIGNED TRIMERIC COILED COIL-VALD | | Descriptor: | COIL-VALD, SULFATE ION | | Authors: | Ogihara, N.L, Weiss, M.S, Degrado, W.F, Eisenberg, D. | | Deposit date: | 1996-08-10 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the designed trimeric coiled coil coil-VaLd: implications for engineering crystals and supramolecular assemblies.
Protein Sci., 6, 1997
|
|
7JRQ
 
 | | Crystallographically Characterized De Novo Designed Mn-Diphenylporphyrin Binding Protein | | Descriptor: | CHLORIDE ION, GLYCEROL, MPP1, ... | | Authors: | Mann, S.I, DeGrado, W.F. | | Deposit date: | 2020-08-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | De Novo Design, Solution Characterization, and Crystallographic Structure of an Abiological Mn-Porphyrin-Binding Protein Capable of Stabilizing a Mn(V) Species.
J.Am.Chem.Soc., 143, 2021
|
|
7JH6
 
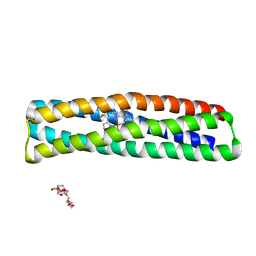 | | De novo designed two-domain di-Zn(II) and porphyrin-binding protein | | Descriptor: | NONAETHYLENE GLYCOL, Two-domain di-Zn(II) and porphyrin-binding protein, ZINC ION, ... | | Authors: | Schmidt, N, Liu, L, DeGrado, W.F. | | Deposit date: | 2020-07-20 | | Release date: | 2020-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Allosteric cooperation in a de novo-designed two-domain protein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2KZ2
 
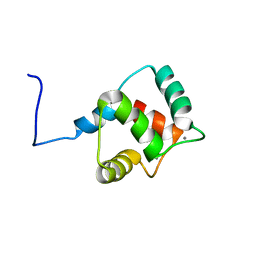 | | Calmodulin, C-terminal domain, F92E mutant | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Korendovych, I, Kulp, D, Wu, Y, Cheng, H, Roder, H, DeGrado, W. | | Deposit date: | 2010-06-10 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design of a switchable eliminase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2LY0
 
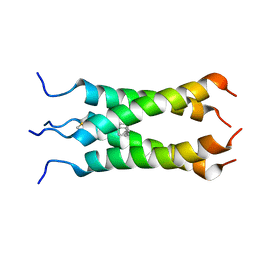 | | Solution NMR structure of the influenza A virus S31N mutant (19-49) in presence of drug M2WJ332 | | Descriptor: | (3S,5S,7S)-N-{[5-(thiophen-2-yl)-1,2-oxazol-3-yl]methyl}tricyclo[3.3.1.1~3,7~]decan-1-aminium, Membrane ion channel M2 | | Authors: | Wu, Y, Wang, J, DeGrado, W. | | Deposit date: | 2012-09-10 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2MUV
 
 | | NOE-based model of the influenza A virus M2 (19-49) bound to drug 11 | | Descriptor: | (3s,5s,7s)-N-[(5-bromothiophen-2-yl)methyl]tricyclo[3.3.1.1~3,7~]decan-1-aminium, Matrix protein 2 | | Authors: | Wu, Y, Wang, J, DeGrado, W. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flipping in the Pore: Discovery of Dual Inhibitors That Bind in Different Orientations to the Wild-Type versus the Amantadine-Resistant S31N Mutant of the Influenza A Virus M2 Proton Channel.
J.Am.Chem.Soc., 136, 2014
|
|
2MUW
 
 | | NOE-based model of the influenza A virus N31S mutant (19-49) bound to drug 11 | | Descriptor: | (3s,5s,7s)-N-[(5-bromothiophen-2-yl)methyl]tricyclo[3.3.1.1~3,7~]decan-1-aminium, Matrix protein 2 | | Authors: | Wu, Y, Wang, J, DeGrado, W. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flipping in the Pore: Discovery of Dual Inhibitors That Bind in Different Orientations to the Wild-Type versus the Amantadine-Resistant S31N Mutant of the Influenza A Virus M2 Proton Channel.
J.Am.Chem.Soc., 136, 2014
|
|
4P6J
 
 | |
