6VBZ
 
 | | Crystal structure of the rat MLKL pseudokinase domain | | Descriptor: | MANGANESE (II) ION, Mixed lineage kinase domain-like pseudokinase | | Authors: | Davies, K.A, Czabotar, P.E. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Distinct pseudokinase domain conformations underlie divergent activation mechanisms among vertebrate MLKL orthologues.
Nat Commun, 11, 2020
|
|
6VC0
 
 | | Crystal structure of the horse MLKL pseudokinase domain | | Descriptor: | GLYCEROL, Mixed lineage kinase domain like pseudokinase | | Authors: | Davies, K.A, Czabotar, P.E. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Distinct pseudokinase domain conformations underlie divergent activation mechanisms among vertebrate MLKL orthologues.
Nat Commun, 11, 2020
|
|
7MX3
 
 | | Crystal structure of human RIPK3 complexed with GSK'843 | | Descriptor: | 1,2-ETHANEDIOL, 3-(1,3-benzothiazol-5-yl)-7-(1,3-dimethyl-1H-pyrazol-5-yl)thieno[3,2-c]pyridin-4-amine, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Davies, K.A, Czabotar, P.E. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Human RIPK3 maintains MLKL in an inactive conformation prior to cell death by necroptosis.
Nat Commun, 12, 2021
|
|
2JK1
 
 | | Crystal structure of the wild-type HupR receiver domain | | Descriptor: | HYDROGENASE TRANSCRIPTIONAL REGULATORY PROTEIN HUPR1, MAGNESIUM ION | | Authors: | Davies, K.M, Lowe, E.D, Venien-Bryan, C, Johnson, L.N. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Hupr Receiver Domain Crystal Structure in its Nonphospho and Inhibitory Phospho States.
J.Mol.Biol., 385, 2009
|
|
4B2Q
 
 | |
2VUH
 
 | |
2VUI
 
 | | Crystal structure of the HupR receiver domain in inhibitory phospho- state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, HYDROGENASE TRANSCRIPTIONAL REGULATORY PROTEIN HUPR1, MAGNESIUM ION | | Authors: | Davies, K.M, Lowe, E.D, Venien-Bryan, C, Johnson, L.N. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Hupr Receiver Domain Crystal Structure in its Nonphospho and Inhibitory Phospho States.
J.Mol.Biol., 385, 2009
|
|
7LTB
 
 | | Crystal Structure of Keratinicyclin B | | Descriptor: | (2~{S},4~{S},5~{R},6~{S})-4-azanyl-5-methoxy-6-methyl-oxan-2-ol, 3-ammonio-2,3,6-trideoxy-alpha-L-arabino-hexopyranose-(1-2)-beta-D-glucopyranose, FORMIC ACID, ... | | Authors: | Davis, K.M, Jeffrey, P.D, Seyedsayamdost, M.R. | | Deposit date: | 2021-02-19 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Potent and specific antibiotic combination therapy against Clostridioides difficile.
Nat.Chem.Biol., 20, 2024
|
|
7LKC
 
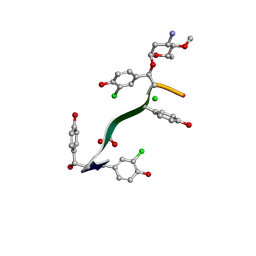 | | Crystal Structure of Keratinimicin A | | Descriptor: | (2~{S},4~{S},5~{R},6~{S})-4-azanyl-5-methoxy-6-methyl-oxan-2-ol, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Davis, K.M, Jeffrey, P.D, Seyedsayamdost, M.R. | | Deposit date: | 2021-02-02 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Potent and specific antibiotic combination therapy against Clostridioides difficile.
Nat.Chem.Biol., 20, 2024
|
|
6EDH
 
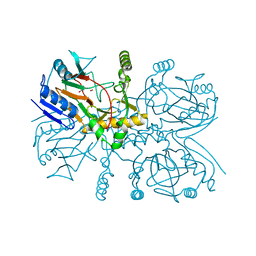 | | Taurine:2OG dioxygenase (TauD) bound to the vanadyl ion, taurine, and succinate | | Descriptor: | 2-AMINOETHANESULFONIC ACID, ACETATE ION, Alpha-ketoglutarate-dependent taurine dioxygenase, ... | | Authors: | Davis, K.M, Altmyer, M, Boal, A.K. | | Deposit date: | 2018-08-09 | | Release date: | 2019-08-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73000407 Å) | | Cite: | Structure of a Ferryl Mimic in the Archetypal Iron(II)- and 2-(Oxo)-glutarate-Dependent Dioxygenase, TauD.
Biochemistry, 58, 2019
|
|
8ELW
 
 | | HRAS R97A Crystal Form 1 T-State | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Davis, K, Johnson, C.W, Mattos, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Allosteric site variants affect GTP hydrolysis on Ras.
Protein Sci., 32, 2023
|
|
5V1Q
 
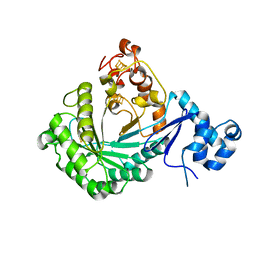 | | Crystal structure of Streptococcus suis SuiB | | Descriptor: | IRON/SULFUR CLUSTER, Radical SAM | | Authors: | Davis, K.M, Bacik, J.P, Ando, N. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V1S
 
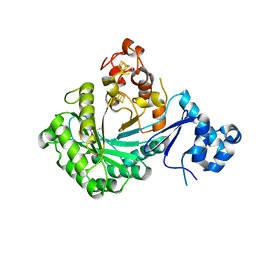 | |
5V1T
 
 | | Crystal structure of Streptococcus suis SuiB bound to precursor peptide SuiA | | Descriptor: | IRON/SULFUR CLUSTER, METHIONINE, Radical SAM, ... | | Authors: | Davis, K.M, Bacik, J.P, Ando, N. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6VP4
 
 | | Ethylene forming enzyme (EFE) in complex with Fe(II), L-arginine, and 2OG | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ... | | Authors: | Davis, K.M, Copeland, R.A, Boal, A.K. | | Deposit date: | 2020-02-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An Iron(IV)-Oxo Intermediate Initiating l-Arginine Oxidation but Not Ethylene Production by the 2-Oxoglutarate-Dependent Oxygenase, Ethylene-Forming Enzyme.
J.Am.Chem.Soc., 143, 2021
|
|
6VP5
 
 | | Ethylene forming enzyme (EFE) D191E variant in complex with Fe(II), L-arginine, and 2OG | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ... | | Authors: | Davis, K.M, Copeland, R.A, Boal, A.K. | | Deposit date: | 2020-02-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | An Iron(IV)-Oxo Intermediate Initiating l-Arginine Oxidation but Not Ethylene Production by the 2-Oxoglutarate-Dependent Oxygenase, Ethylene-Forming Enzyme.
J.Am.Chem.Soc., 143, 2021
|
|
6NBX
 
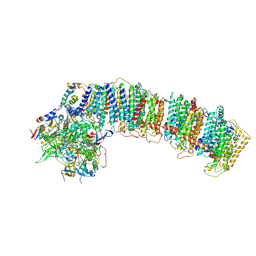 | | T.elongatus NDH (data-set 2) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 1, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
6NBQ
 
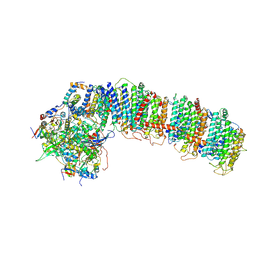 | | T.elongatus NDH (data-set 1) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 2, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
6NBY
 
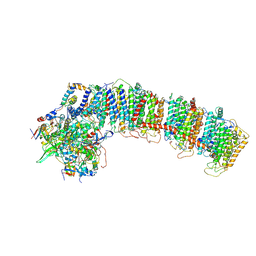 | | T.elongatus NDH (composite model) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 1, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-27 | | Last modified: | 2020-04-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
8SLZ
 
 | |
6OWG
 
 | | Structure of a synthetic beta-carboxysome shell, T=4 | | Descriptor: | Ethanolamine utilization protein EutN/carboxysome structural protein Ccml, Microcompartments protein | | Authors: | Sutter, M, Laughlin, T.G, Davies, K.M, Kerfeld, C.A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of a Syntheticbeta-Carboxysome Shell.
Plant Physiol., 181, 2019
|
|
7MON
 
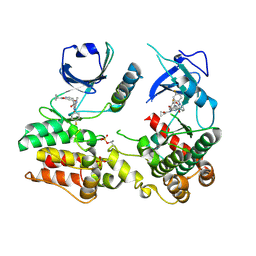 | | Structure of human RIPK3-MLKL complex | | Descriptor: | Mixed lineage kinase domain-like protein, N-[4-({2-[(cyclopropanecarbonyl)amino]pyridin-4-yl}oxy)-3-fluorophenyl]-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Meng, Y, Davies, K.A, Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2021-05-03 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Human RIPK3 maintains MLKL in an inactive conformation prior to cell death by necroptosis.
Nat Commun, 12, 2021
|
|
6GNH
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and an Azepanyl Phenyl Benzylsulphonamide Ligand | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, methyl 4-(azepan-1-yl)-3-[(4-methoxyphenyl)sulfonylamino]benzoate | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6GNV
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and a isopropyl methyl indole aryl sulphonamide ligand | | Descriptor: | GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, ... | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6GNS
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and an Azepanyl Phenyl Benzylsulphonamide Ligand | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, methyl 4-(azepan-1-yl)-3-[[4-[4-(1-methylpiperidin-4-yl)butyl]phenyl]sulfonylamino]benzoate | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
