6AUA
 
 | |
5MW6
 
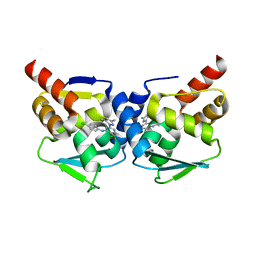 | | Crystal structure of the BCL6 BTB-domain with compound 1 | | Descriptor: | 5-chloranyl-~{N}-(4-chlorophenyl)-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-amine, B-cell lymphoma 6 protein | | Authors: | Davies, D.R, Kessler, D. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
8ELS
 
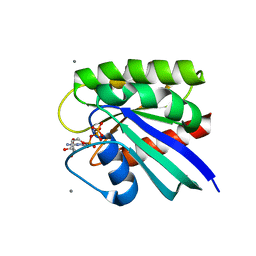 | | HRAS R97A Crystal Form 1 R-state | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Davis, D, Johnson, C.W, Mattos, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.267 Å) | | Cite: | Allosteric site variants affect GTP hydrolysis on Ras.
Protein Sci., 32, 2023
|
|
7UMP
 
 | | CRYSTAL STRUCTURE OF PHD2 CATALYTIC DOMAIN (CID 7465) IN COMPLEX WITH AKB-6548 AT 1.8 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, Egl nine homolog 1, FE (II) ION, ... | | Authors: | Davie, D.R, Abendroth, J, Boyd, J. | | Deposit date: | 2022-04-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Preclinical Characterization of Vadadustat (AKB-6548), an Oral Small Molecule Hypoxia-Inducible Factor Prolyl-4-Hydroxylase Inhibitor, for the Potential Treatment of Renal Anemia.
J.Pharmacol.Exp.Ther., 383, 2022
|
|
1GK5
 
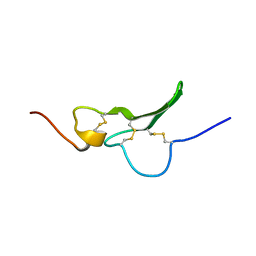 | | Solution Structure the mEGF/TGFalpha44-50 chimeric growth factor | | Descriptor: | Pro-epidermal growth factor,Protransforming growth factor alpha | | Authors: | Chamberlin, S.G, Brennan, L, Puddicombe, S.M, Davies, D.E, Turner, D.L. | | Deposit date: | 2001-08-08 | | Release date: | 2002-08-08 | | Last modified: | 2018-03-28 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Megf/Tgfalpha44-50 Chimeric Growth Factor.
Eur.J.Biochem., 268, 2001
|
|
4NPC
 
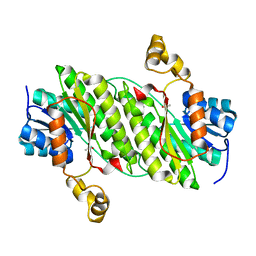 | | Crystal Structure of an Oxidoreductase, Short-Chain Dehydrogenase/Reductase Family Protein from Brucella suis | | Descriptor: | ACETATE ION, Sorbitol dehydrogenase | | Authors: | Dranow, D.M, Davies, D.R, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of an Oxidoreductase, Short-Chain Dehydrogenase/Reductase Family Protein from Brucella suis
To be Published
|
|
1TTQ
 
 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) IN THE PRESENCE OF POTASSIUM AT ROOM TEMPERATURE | | Descriptor: | POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE | | Authors: | Rhee, S, Parris, K, Ahmed, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase alpha2beta2 complex.
Biochemistry, 35, 1996
|
|
1TTP
 
 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) IN THE PRESENCE OF CESIUM, ROOM TEMPERATURE | | Descriptor: | CESIUM ION, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE | | Authors: | Rhee, S, Parris, K, Ahmed, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase alpha2beta2 complex.
Biochemistry, 35, 1996
|
|
1UBS
 
 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) WITH A MUTATION OF LYS 87->THR IN THE B SUBUNIT AND IN THE PRESENCE OF LIGAND L-SERINE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE, SODIUM ION, ... | | Authors: | Rhee, S, Parris, K, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-12-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
1MCP
 
 | | PHOSPHOCHOLINE BINDING IMMUNOGLOBULIN FAB MC/PC603. AN X-RAY DIFFRACTION STUDY AT 2.7 ANGSTROMS | | Descriptor: | IGA-KAPPA MCPC603 FAB (HEAVY CHAIN), IGA-KAPPA MCPC603 FAB (LIGHT CHAIN), SULFATE ION | | Authors: | Satow, Y, Cohen, G.H, Padlan, E.A, Davies, D.R. | | Deposit date: | 1984-07-09 | | Release date: | 1985-01-02 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Phosphocholine binding immunoglobulin Fab McPC603. An X-ray diffraction study at 2.7 A.
J.Mol.Biol., 190, 1986
|
|
1AER
 
 | | DOMAIN III OF PSEUDOMONAS AERUGINOSA EXOTOXIN COMPLEXED WITH BETA-TAD | | Descriptor: | 2-(1,5-DIDEOXYRIBOSE)-4-AMIDO-THIAZOLE, ADENOSINE MONOPHOSPHATE, BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, M, Dyda, F, Benhar, I, Pastan, I, Davies, D.R. | | Deposit date: | 1995-12-11 | | Release date: | 1996-06-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of Pseudomonas exotoxin A complexed with a nicotinamide adenine dinucleotide analog: implications for the activation process and for ADP ribosylation
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1A30
 
 | | HIV-1 PROTEASE COMPLEXED WITH A TRIPEPTIDE INHIBITOR | | Descriptor: | HIV-1 PROTEASE, TRIPEPTIDE GLU-ASP-LEU | | Authors: | Louis, J.M, Dyda, F, Nashed, N.T, Kimmel, A.R, Davies, D.R. | | Deposit date: | 1998-01-27 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophilic peptides derived from the transframe region of Gag-Pol inhibit the HIV-1 protease.
Biochemistry, 37, 1998
|
|
1A5A
 
 | | CRYO-CRYSTALLOGRAPHY OF A TRUE SUBSTRATE, INDOLE-3-GLYCEROL PHOSPHATE, BOUND TO A MUTANT (ALPHAD60N) TRYPTOPHAN SYNTHASE ALPHA2BETA2 COMPLEX REVEALS THE CORRECT ORIENTATION OF ACTIVE SITE ALPHA GLU 49 | | Descriptor: | POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE (ALPHA CHAIN), ... | | Authors: | Rhee, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1998-02-12 | | Release date: | 1998-05-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryo-crystallography of a true substrate, indole-3-glycerol phosphate, bound to a mutant (alphaD60N) tryptophan synthase alpha2beta2 complex reveals the correct orientation of active site alphaGlu49.
J.Biol.Chem., 273, 1998
|
|
1A5B
 
 | | CRYO-CRYSTALLOGRAPHY OF A TRUE SUBSTRATE, INDOLE-3-GLYCEROL PHOSPHATE, BOUND TO A MUTANT (ALPHA D60N) TRYPTOPHAN SYNTHASE ALPHA2BETA2 COMPLEX REVEALS THE CORRECT ORIENTATION OF ACTIVE SITE ALPHA GLU 49 | | Descriptor: | INDOLE-3-GLYCEROL PHOSPHATE, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Rhee, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1998-02-12 | | Release date: | 1998-05-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-crystallography of a true substrate, indole-3-glycerol phosphate, bound to a mutant (alphaD60N) tryptophan synthase alpha2beta2 complex reveals the correct orientation of active site alphaGlu49.
J.Biol.Chem., 273, 1998
|
|
2APR
 
 | |
5VPV
 
 | |
4APR
 
 | |
6APR
 
 | |
5APR
 
 | |
2A0Z
 
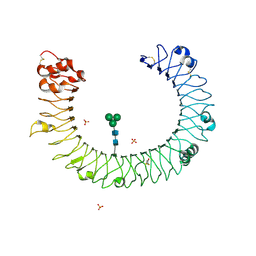 | | The molecular structure of toll-like receptor 3 ligand binding domain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bell, J.K, Botos, I, Hall, P.R, Askins, J, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2005-06-17 | | Release date: | 2005-08-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular structure of the Toll-like receptor 3 ligand-binding domain
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3HFM
 
 | |
7AJR
 
 | | Virtual screening approach leading to the identification of a novel and tractable series of Pseudomonas aeruginosa elastase inhibitors | | Descriptor: | 2-[2-(1,3-benzothiazol-2-ylmethylcarbamoyl)-1,3-dihydroinden-2-yl]ethanoic acid, Keratinase KP2, SULFATE ION, ... | | Authors: | Leiris, S, Davies, D.T, Sprinsky, N, Castandet, J, Behria, L, Bodnarchuk, M.S, Sutton, J.M, Mullins, T.M.G, Jones, M.W, Forrest, A.K, Pallin, T.D, Karunakar, P, Martha, S.K, Parusharamulu, B, Ramula, R, Kotha, V, Pottabathini, N, Pothukanuri, S, Lemonnier, M, Everett, M. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual Screening Approach to Identifying a Novel and Tractable Series of Pseudomonas aeruginosa Elastase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
1ITG
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HIV-1 INTEGRASE: SIMILARITY TO OTHER POLYNUCLEOTIDYL TRANSFERASES | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Dyda, F, Hickman, A.B, Jenkins, T.M, Engelman, A, Craigie, R, Davies, D.R. | | Deposit date: | 1994-11-21 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases.
Science, 266, 1994
|
|
1ZYM
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI | | Descriptor: | ENZYME I | | Authors: | Liao, D.-I, Davies, D.R. | | Deposit date: | 1996-05-21 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The first step in sugar transport: crystal structure of the amino terminal domain of enzyme I of the E. coli PEP: sugar phosphotransferase system and a model of the phosphotransfer complex with HPr.
Structure, 4, 1996
|
|
2MCP
 
 | |
