6NIB
 
 | | Crystal Structure of Medicago truncatula Agmatine Iminohydrolase (Deiminase) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Porphyromonas-type peptidyl-arginine deiminase, ... | | Authors: | Sekula, B, Dauter, Z. | | Deposit date: | 2018-12-27 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Study of Agmatine Iminohydrolase FromMedicago truncatula, the Second Enzyme of the Agmatine Route of Putrescine Biosynthesis in Plants.
Front Plant Sci, 10, 2019
|
|
6NIC
 
 | | Crystal Structure of Medicago truncatula Agmatine Iminohydrolase (Deiminase) in Complex with 6-aminohexanamide | | Descriptor: | 1,2-ETHANEDIOL, 6-aminohexanamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Dauter, Z. | | Deposit date: | 2018-12-27 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Study of Agmatine Iminohydrolase FromMedicago truncatula, the Second Enzyme of the Agmatine Route of Putrescine Biosynthesis in Plants.
Front Plant Sci, 10, 2019
|
|
6O65
 
 | |
1OR0
 
 | | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation | | Descriptor: | 1,2-ETHANEDIOL, Glutaryl 7-Aminocephalosporanic Acid Acylase, glutaryl acylase | | Authors: | Kim, J.K, Yang, I.S, Rhee, S, Dauter, Z, Lee, Y.S, Park, S.S, Kim, K.H. | | Deposit date: | 2003-03-11 | | Release date: | 2004-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation
Biochemistry, 42, 2003
|
|
1OTG
 
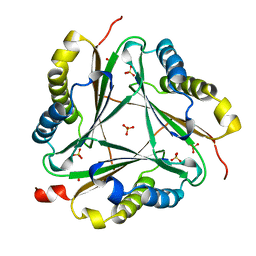 | | 5-CARBOXYMETHYL-2-HYDROXYMUCONATE ISOMERASE | | Descriptor: | 5-CARBOXYMETHYL-2-HYDROXYMUCONATE ISOMERASE, SULFATE ION | | Authors: | Subramanya, H.S, Roper, D.I, Dauter, Z, Dodson, E.J, Davies, G.J, Wilson, K.S, Wigley, D.B. | | Deposit date: | 1995-11-09 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzymatic ketonization of 2-hydroxymuconate: specificity and mechanism investigated by the crystal structures of two isomerases.
Biochemistry, 35, 1996
|
|
1OPO
 
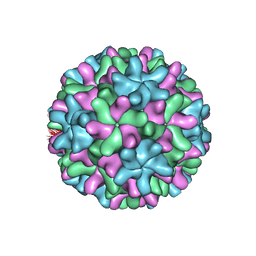 | | THE STRUCTURE OF CARNATION MOTTLE VIRUS | | Descriptor: | CALCIUM ION, Coat protein, SULFATE ION | | Authors: | Morgunova, E, Dauter, Z, Fry, E, Stuart, D, Stel'mashchuk, V, Mikhailov, A.M, Wilson, K.S, Vainshtein, B.K. | | Deposit date: | 2003-03-06 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The atomic structure of Carnation Mottle Virus capsid protein
Febs Lett., 338, 1994
|
|
1OTF
 
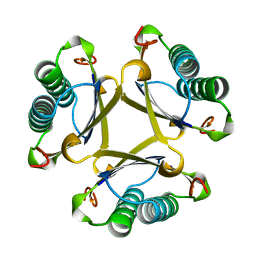 | | 4-OXALOCROTONATE TAUTOMERASE-TRICLINIC CRYSTAL FORM | | Descriptor: | 4-OXALOCROTONATE TAUTOMERASE | | Authors: | Subramanya, H.S, Roper, D.I, Dauter, Z, Dodson, E.J, Davies, G.J, Wilson, K.S, Wigley, D.B. | | Deposit date: | 1995-11-09 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enzymatic ketonization of 2-hydroxymuconate: specificity and mechanism investigated by the crystal structures of two isomerases.
Biochemistry, 35, 1996
|
|
1PAZ
 
 | |
1PNE
 
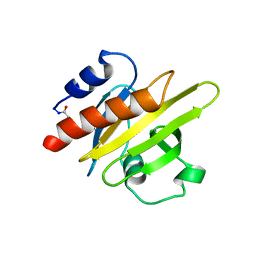 | | CRYSTALLIZATION AND STRUCTURE DETERMINATION OF BOVINE PROFILIN AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | PROFILIN | | Authors: | Cedergren-Zeppezauer, E.S, Goonesekere, N.C.W, Rozycki, M.D, Myslik, J.C, Dauter, Z, Lindberg, U, Schutt, C.E. | | Deposit date: | 1995-05-05 | | Release date: | 1995-07-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and structure determination of bovine profilin at 2.0 A resolution.
J.Mol.Biol., 240, 1994
|
|
1PY3
 
 | | Crystal structure of Ribonuclease Sa2 | | Descriptor: | SULFATE ION, ribonuclease | | Authors: | Sevcik, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 2003-07-08 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure reveals two alternative conformations in the active site of ribonuclease Sa2.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PYL
 
 | | Crystal structure of Ribonuclease Sa2 | | Descriptor: | SULFATE ION, ribonuclease | | Authors: | Sevcik, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 2003-07-09 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | Crystal structure reveals two alternative conformations in the active site of ribonuclease Sa2.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PSP
 
 | | PANCREATIC SPASMOLYTIC POLYPEPTIDE: FIRST THREE-DIMENSIONAL STRUCTURE OF A MEMBER OF THE MAMMALIAN TREFOIL FAMILY OF PEPTIDES | | Descriptor: | PANCREATIC SPASMOLYTIC POLYPEPTIDE | | Authors: | Gajhede, M, Petersen, T.N, Henriksen, A, Petersen, J.F.W, Dauter, Z, Wilson, K.S, Thim, L. | | Deposit date: | 1994-01-05 | | Release date: | 1994-04-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pancreatic spasmolytic polypeptide: first three-dimensional structure of a member of the mammalian trefoil family of peptides.
Structure, 1, 1993
|
|
1QBB
 
 | | BACTERIAL CHITOBIASE COMPLEXED WITH CHITOBIOSE (DINAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-07 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1QBA
 
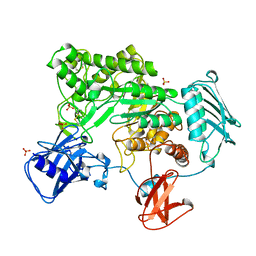 | | BACTERIAL CHITOBIASE, GLYCOSYL HYDROLASE FAMILY 20 | | Descriptor: | CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1RRE
 
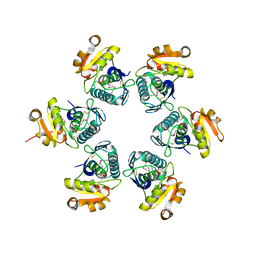 | | Crystal structure of E.coli Lon proteolytic domain | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Rasulova, F, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1RR9
 
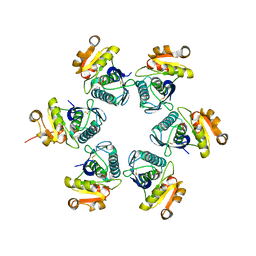 | | Catalytic domain of E.coli Lon protease | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2003-12-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1SCR
 
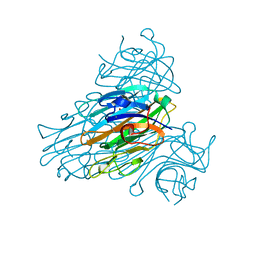 | | HIGH-RESOLUTION STRUCTURES OF SINGLE-METAL-SUBSTITUTED CONCANAVALIN A: THE CO,CA-PROTEIN AT 1.6 ANGSTROMS AND THE NI,CA-PROTEIN AT 2.0 ANGSTROMS | | Descriptor: | CALCIUM ION, CONCANAVALIN A, NICKEL (II) ION | | Authors: | Emmerich, C, Helliwell, J.R, Redshaw, M, Naismith, J.H, Harrop, S.J, Raftery, J, Kalb, A.J, Yariv, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 1993-12-06 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of single-metal-substituted concanavalin A: the Co,Ca-protein at 1.6 A and the Ni,Ca-protein at 2.0 A.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1SCS
 
 | | HIGH-RESOLUTION STRUCTURES OF SINGLE-METAL-SUBSTITUTED CONCANAVALIN A: THE CO,CA-PROTEIN AT 1.6 ANGSTROMS AND THE NI,CA-PROTEIN AT 2.0 ANGSTROMS | | Descriptor: | CALCIUM ION, COBALT (II) ION, CONCANAVALIN A | | Authors: | Emmerich, C, Helliwell, J.R, Redshaw, M, Naismith, J.H, Harrop, S.J, Raftery, J, Kalb, A.J, Yariv, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 1993-12-06 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of single-metal-substituted concanavalin A: the Co,Ca-protein at 1.6 A and the Ni,Ca-protein at 2.0 A.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1KLL
 
 | | Molecular basis of mitomycin C resictance in streptomyces: Crystal structures of the MRD protein with and without a drug derivative | | Descriptor: | 1,2-CIS-1-HYDROXY-2,7-DIAMINO-MITOSENE, mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-12 | | Release date: | 2002-07-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
1KMT
 
 | | Crystal structure of RhoGDI Glu(154,155)Ala mutant | | Descriptor: | Rho GDP-dissociation inhibitor 1 | | Authors: | Mateja, A, Devedjiev, Y, Krowarsh, D, Longenecker, K, Dauter, Z, Otlewski, J, Derewenda, Z.S. | | Deposit date: | 2001-12-17 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The impact of Glu-->Ala and Glu-->Asp mutations on the crystallization properties of RhoGDI: the structure of RhoGDI at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KMZ
 
 | | MOLECULAR BASIS OF MITOMYCIN C RESICTANCE IN STREPTOMYCES: CRYSTAL STRUCTURES OF THE MRD PROTEIN WITH AND WITHOUT A DRUG DERIVATIVE | | Descriptor: | mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
1LQV
 
 | | Crystal structure of the Endothelial protein C receptor with phospholipid in the groove in complex with Gla domain of protein C. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endothelial protein C receptor, ... | | Authors: | Oganesyan, V, Oganesyan, N, Terzyan, S, Dongfeng, Q, Dauter, Z, Esmon, N.L, Esmon, C.T. | | Deposit date: | 2002-05-13 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the endothelial protein C receptor and a bound phospholipid.
J.Biol.Chem., 277, 2002
|
|
1LNI
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS AT ATOMIC RESOLUTION (1.0 A) | | Descriptor: | GLYCEROL, GUANYL-SPECIFIC RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Lamzin, V.S, Dauter, Z, Wilson, K.S. | | Deposit date: | 2002-05-03 | | Release date: | 2002-07-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution data reveal flexibility in the structure of RNase Sa.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1L8J
 
 | | Crystal Structure of the Endothelial Protein C Receptor and Bound Phospholipid Molecule | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endothelial protein C receptor, ... | | Authors: | Oganesyan, V, Oganesyan, N, Terzyan, S, Dongfeng, Q, Dauter, Z, Esmon, N.L, Esmon, C.T. | | Deposit date: | 2002-03-20 | | Release date: | 2002-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the endothelial protein C receptor and a bound phospholipid.
J.Biol.Chem., 277, 2002
|
|
1MJD
 
 | | Structure of N-terminal domain of human doublecortin | | Descriptor: | DOUBLECORTIN | | Authors: | Kim, M.H, Cierpicki, T, Derewenda, U, Krowarsch, D, Feng, Y, Devedjiev, Y, Dauter, Z, Walsh, C.A, Otlewski, J, Bushweller, J.H, Derewenda, Z.S. | | Deposit date: | 2002-08-27 | | Release date: | 2003-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The DCX-domain Tandems of Doublecortin and Doublecortin-like Kinase
Nat.Struct.Biol., 10, 2003
|
|
