8JI1
 
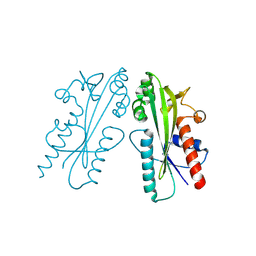 | | Crystal structure of Ham1 from Plasmodium falciparum | | Descriptor: | Inosine triphosphate pyrophosphatase | | Authors: | Pramanik, A, Datta, S. | | Deposit date: | 2023-05-25 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-function analysis of nucleotide housekeeping protein HAM1 from human malaria parasite Plasmodium falciparum.
Febs J., 2024
|
|
2A0I
 
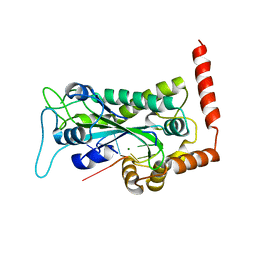 | | F Factor TraI Relaxase Domain bound to F oriT Single-stranded DNA | | Descriptor: | F plasmid single-stranded oriT DNA, IMIDAZOLE, MAGNESIUM ION, ... | | Authors: | Larkin, C, Datta, S, Harley, M.J, Anderson, B.J, Ebie, A, Hargreaves, V, Schildbach, J.F. | | Deposit date: | 2005-06-16 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the f factor relaxase.
Structure, 13, 2005
|
|
8HUC
 
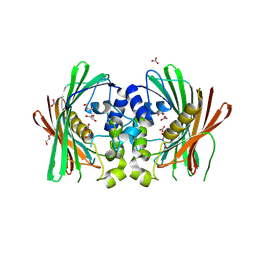 | | Crystal structure of PaIch (Pec1) | | Descriptor: | GLYCEROL, MaoC_dehydrat_N domain-containing protein, NITRATE ION | | Authors: | Pramanik, A, Datta, S. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structural and functional insights of itaconyl-CoA hydratase from Pseudomonas aeruginosa highlight a novel N-terminal hotdog fold.
Febs Lett., 598, 2024
|
|
8IX9
 
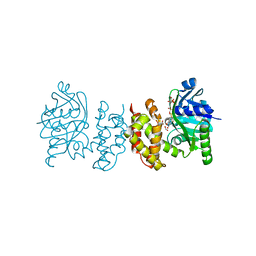 | | Pseudomoans Aerugiona Wildtype Ketopantoate Reductase with NADPH | | Descriptor: | 2-dehydropantoate 2-reductase, FORMIC ACID, GLYCEROL, ... | | Authors: | Choudhury, G.B, Datta, S. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Implication of Molecular Constraints Facilitating the Functional Evolution of Pseudomonas aeruginosa KPR2 into a Versatile alpha-Keto-Acid Reductase.
Biochemistry, 63, 2024
|
|
8IWG
 
 | |
8IXH
 
 | |
8IWQ
 
 | |
8IXM
 
 | |
7XXN
 
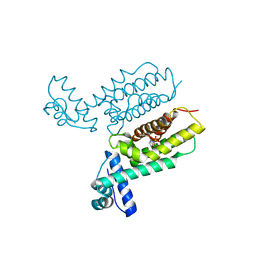 | | HapR Quadruple mutant, bound to Qstatin | | Descriptor: | 1-(5-bromanylthiophen-2-yl)sulfonylpyrazole, GLYCEROL, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XY0
 
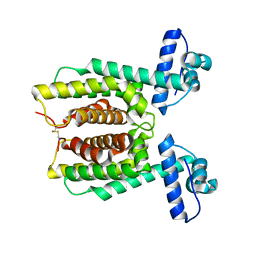 | | HapR Double mutant Y76F, F171C | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-31 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7Y4J
 
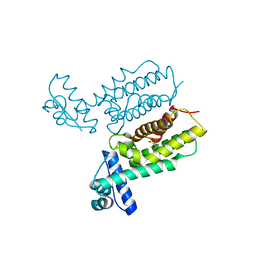 | | HapR_Triple mutant Y76F, L97I, F171C | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXS
 
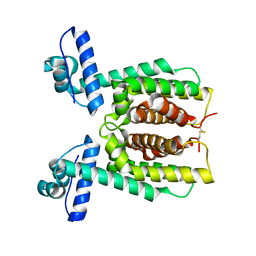 | | HapR mutant I141V | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XYI
 
 | | HapR Quadruple mutant Y76F, L97I, I141V, F171C | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXO
 
 | | HapR Native in CHES buffer pH 9.5 | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XY5
 
 | | HapR_Double Mutant with CHES buffer | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-31 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXT
 
 | | HapR Quadruple mutant, bound to IMTVC-212 | | Descriptor: | 1-((5-phenylthiophen-2-yl)sulfonyl)-1H-pyrazole, DIMETHYL SULFOXIDE, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
8I2K
 
 | |
3X1M
 
 | |
3X1K
 
 | |
3X1J
 
 | |
3NBJ
 
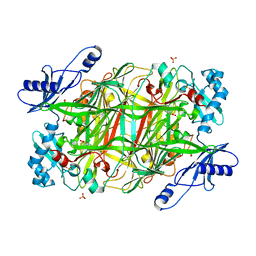 | | Crystal Structure of Y305F mutant of the copper amine oxidase from Hansenula polymorpha expressed in yeast | | Descriptor: | COPPER (II) ION, PHOSPHATE ION, Peroxisomal primary amine oxidase | | Authors: | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2010-06-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
3NBB
 
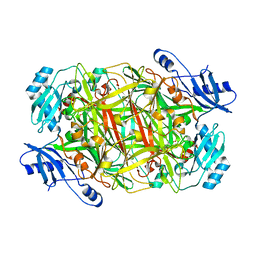 | | Crystal structure of mutant Y305F expressed in E. coli in the copper amine oxidase from hansenula polymorpha | | Descriptor: | COPPER (II) ION, Peroxisomal primary amine oxidase | | Authors: | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2010-06-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
3N9H
 
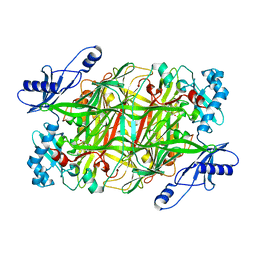 | | Crystal Structural of mutant Y305A in the copper amine oxidase from hansenula polymorpha | | Descriptor: | COPPER (II) ION, Peroxisomal primary amine oxidase | | Authors: | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2010-05-30 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
2ODW
 
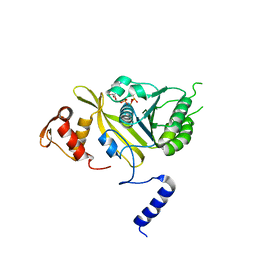 | | MSrecA-ATP-GAMA-S complex | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-12-27 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
5XCH
 
 | | Crystal structure of Wild type Vps29 complexed with Zn+2 from Entamoeba histolytica | | Descriptor: | Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Srivastava, V.K, Yadav, R, Tomar, P, Gourinath, S, Datta, S. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and thermodynamic characterization of metal binding in Vps29 from Entamoeba histolytica: implication in retromer function.
Mol. Microbiol., 106, 2017
|
|
