9J4Y
 
 | |
9J4Z
 
 | |
9J50
 
 | |
9J2K
 
 | |
7DFY
 
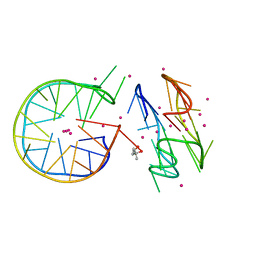 | | Novel motif for left-handed G-quadruplex formation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2xMotif2, POTASSIUM ION, ... | | Authors: | Das, P, Winnerdy, F.R, Maity, A, Mechulam, Y, Phan, A.T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A novel minimal motif for left-handed G-quadruplex formation.
Chem.Commun.(Camb.), 57, 2021
|
|
7D5D
 
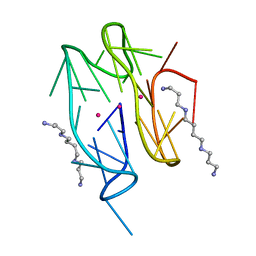 | | Left-handed G-quadruplex containing one bulge | | Descriptor: | 1xBulge-LHG4motif, POTASSIUM ION, SPERMINE | | Authors: | Das, P, Ngo, K.H, Winnerdy, F.R, Maity, A, Bakalar, B, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Bulges in left-handed G-quadruplexes.
Nucleic Acids Res., 49, 2021
|
|
7D5E
 
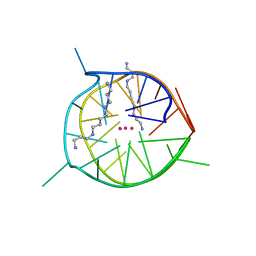 | | Left-handed G-quadruplex containing two bulges | | Descriptor: | 2xBulge-LHG4motif, POTASSIUM ION, SODIUM ION, ... | | Authors: | Das, P, Maity, A, Ngo, K.H, Winnerdy, F.R, Bakalar, B, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | Bulges in left-handed G-quadruplexes.
Nucleic Acids Res., 49, 2021
|
|
7XTF
 
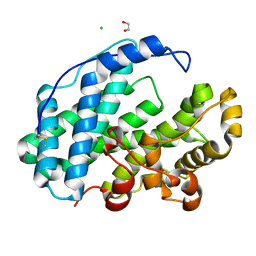 | |
7XTE
 
 | |
7WXM
 
 | |
7WXR
 
 | |
7WXN
 
 | |
7WXP
 
 | |
7WXL
 
 | |
7WXO
 
 | |
7WXQ
 
 | |
7WXK
 
 | |
7WXJ
 
 | |
5Z19
 
 | |
5Z1A
 
 | |
6JZ2
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with uronic isofagomine at 1.3 Angstroms resolution | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6JZ4
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro-d-lactam | | Descriptor: | (2S,3R,4S,5R)-3,4,5-trihydroxy-6-oxopiperidine-2-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6JZ1
 
 | |
5Z18
 
 | |
5Z1B
 
 | |
