4Y6K
 
 | | Complex structure of presenilin homologue PSH bound to an inhibitor | | Descriptor: | N-{(2R,4S,5S)-2-benzyl-5-[(tert-butoxycarbonyl)amino]-4-hydroxy-6-phenylhexanoyl}-L-leucyl-L-phenylalaninamide, Uncharacterized protein PSH | | Authors: | Dang, S, Wu, S, Wang, J, Shi, Y. | | Deposit date: | 2015-02-13 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.855 Å) | | Cite: | Cleavage of amyloid precursor protein by an archaeal presenilin homologue PSH
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6BGJ
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in LMNG | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, L, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
6BGI
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, J, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
8KG6
 
 | | Yeast replisome in state I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
6O20
 
 | | Cryo-EM structure of TRPV5 with calmodulin bound | | Descriptor: | CALCIUM ION, Calmodulin, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Dang, S, van Goor, M.K, Asarnow, D, Wang, Y, Julius, D, Cheng, Y, van der Wijst, J. | | Deposit date: | 2019-02-22 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into TRPV5 channel function and modulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O1N
 
 | | Cryo-EM structure of TRPV5 (1-660) in nanodisc | | Descriptor: | Transient receptor potential cation channel subfamily V member 5 | | Authors: | Dang, S, van Goor, M.K, Asarnow, D, Wang, Y, Julius, D, Cheng, Y, van der Wijst, J. | | Deposit date: | 2019-02-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into TRPV5 channel function and modulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O1U
 
 | | Cryo-EM structure of TRPV5 W583A in nanodisc | | Descriptor: | Transient receptor potential cation channel subfamily V member 5 | | Authors: | Dang, S, van Goor, M.K, Asarnow, D, Wang, Y, Julius, D, Cheng, Y, van der Wijst, J. | | Deposit date: | 2019-02-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight into TRPV5 channel function and modulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O1P
 
 | | Cryo-EM structure of full length TRPV5 in nanodisc | | Descriptor: | Transient receptor potential cation channel subfamily V member 5 | | Authors: | Dang, S, van Goor, M.K, Asarnow, D, Wang, Y, Julius, D, Cheng, Y, van der Wijst, J. | | Deposit date: | 2019-02-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insight into TRPV5 channel function and modulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8KG8
 
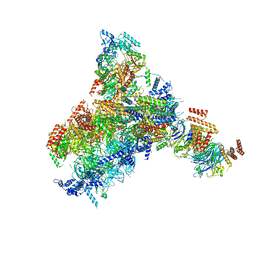 | | Yeast replisome in state II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8KG9
 
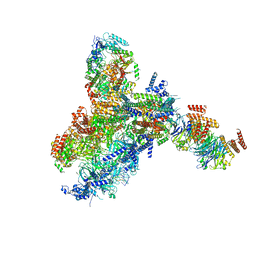 | | Yeast replisome in state III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8W7S
 
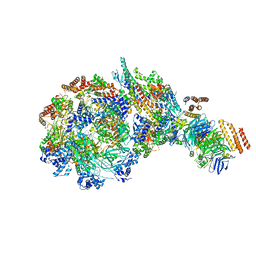 | | Yeast replisome in state IV | | Descriptor: | Cell division control protein 45, DNA (71-mer), DNA polymerase alpha-binding protein, ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D. | | Deposit date: | 2023-08-31 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (7.39 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8W7M
 
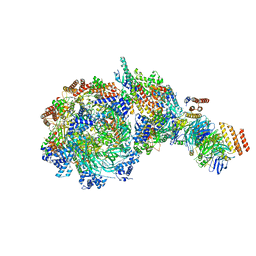 | | Yeast replisome in state V | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (71-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
7YKJ
 
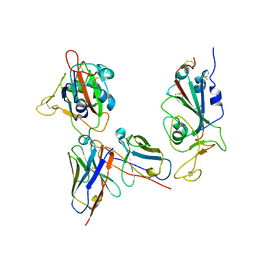 | | Omicron RBDs bound with P3E6 Fab (one up and one down) | | Descriptor: | P3E6 heavy chain, P3E6 light chain, Spike glycoprotein | | Authors: | Tang, B, Dang, S. | | Deposit date: | 2022-07-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into broadly neutralizing antibodies elicited by hybrid immunity against SARS-CoV-2.
Emerg Microbes Infect, 12, 2023
|
|
6P49
 
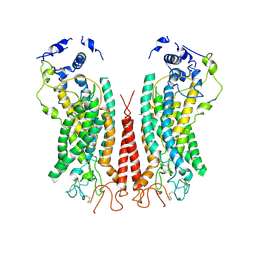 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl2 | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P46
 
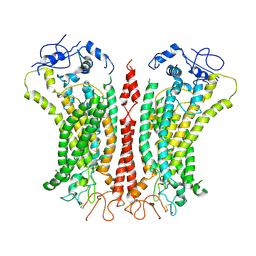 | | Cryo-EM structure of TMEM16F in digitonin with calcium bound | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P47
 
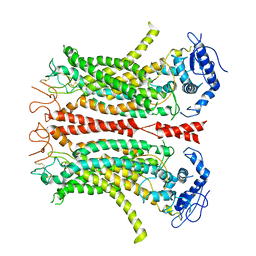 | | Cryo-EM structure of TMEM16F in digitonin without calcium | | Descriptor: | Anoctamin-6 | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P48
 
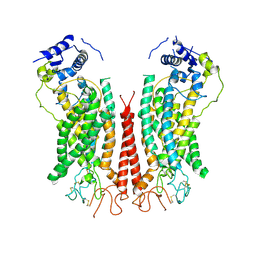 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl1 | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
8I4L
 
 | | Capsid structure of the Cyanophage P-SCSP1u | | Descriptor: | The capsid protein(gp 19) of P-SCSP1u | | Authors: | Liu, H, Dang, S. | | Deposit date: | 2023-01-19 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Cryo-EM structure of cyanophage P-SCSP1u offers insights into DNA gating and evolution of T7-like viruses.
Nat Commun, 14, 2023
|
|
8I4M
 
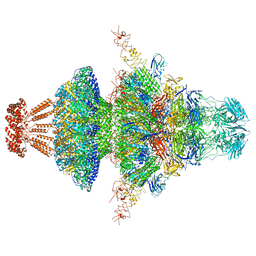 | | Portal-tail complex structure of the Cyanophage P-SCSP1u | | Descriptor: | Adaptor protein(gp22) of the cyanophage P-SCSP1u, Fiber protein(gp 28) of the cyanophage P-SCSP1u, Nozzle protein(gp 23) of the cyanophage P-SCSP1u, ... | | Authors: | Liu, H, Dang, S. | | Deposit date: | 2023-01-19 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Cryo-EM structure of cyanophage P-SCSP1u offers insights into DNA gating and evolution of T7-like viruses.
Nat Commun, 14, 2023
|
|
7XH8
 
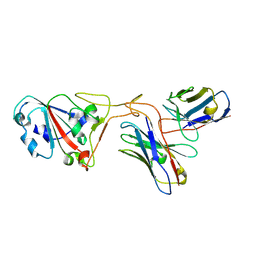 | | The structure of ZCB11 Fab against SARS-CoV-2 Omicron Spike | | Descriptor: | Spike glycoprotein, The heavy chain of ZCB11 antibody, The light chain of ZCB11 antibody | | Authors: | Hang, L, Dang, S. | | Deposit date: | 2022-04-07 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | A broadly neutralizing antibody protects Syrian hamsters against SARS-CoV-2 Omicron challenge.
Nat Commun, 13, 2022
|
|
8K0B
 
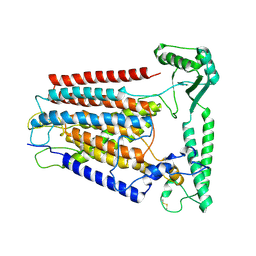 | | Cryo-EM structure of TMEM63C | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Qin, Y, Yu, D, Dong, J, Dang, S. | | Deposit date: | 2023-07-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM structure of TMEM63C suggests it functions as a monomer.
Nat Commun, 14, 2023
|
|
8K19
 
 | |
4HYD
 
 | | Structure of a presenilin family intramembrane aspartate protease in C2221 space group | | Descriptor: | Putative uncharacterized protein | | Authors: | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
4HYG
 
 | | Structure of a presenilin family intramembrane aspartate protease in C222 space group | | Descriptor: | Putative uncharacterized protein | | Authors: | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
4HYC
 
 | | Structure of a presenilin family intramembrane aspartate protease in P2 space group | | Descriptor: | Putative uncharacterized protein | | Authors: | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
