1GPJ
 
 | | Glutamyl-tRNA Reductase from Methanopyrus kandleri | | Descriptor: | (2R,3R,4S,5S)-4-AMINO-2-[6-(DIMETHYLAMINO)-9H-PURIN-9-YL]-5-(HYDROXYMETHYL)TETRAHYDRO-3-FURANOL, CITRIC ACID, GLUTAMIC ACID, ... | | Authors: | Moser, J, Schubert, W.-D, Beier, V, Bringemeier, I, Jahn, D, Heinz, D.W. | | Deposit date: | 2001-11-05 | | Release date: | 2002-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis.
EMBO J., 20, 2001
|
|
7CXO
 
 | | Crystal structure of Arabinose isomerase from hybrid AI10 | | Descriptor: | GLYCEROL, L-arabinose isomerase, MANGANESE (II) ION | | Authors: | Hoang, N.K.Q, Dhanasingh, I, Cao, T.P, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-09-02 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic hybrid AI10
To Be Published
|
|
7CYY
 
 | | Crystal structure of Arabinose isomerase from hybrid AI8 with Adonitol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, D-ribitol, L-arabinose isomerase, ... | | Authors: | Hoang, N.K.Q, Dhanasingh, I, Cao, T.P, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-09-05 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic hybrid AI8 with Adonitol
To Be Published
|
|
6H8E
 
 | | Truncated derivative of the C-terminal domain of the TssA component of the type VI secretion system from Burkholderia cenocepacia | | Descriptor: | Type VI secretion protein ImpA | | Authors: | Dix, S.R, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Brooker, T.A, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | Deposit date: | 2018-08-02 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
1O7X
 
 | | Citrate synthase from Sulfolobus solfataricus | | Descriptor: | CITRATE SYNTHASE | | Authors: | Bell, G.S, Russell, R.J.M, Connaris, H, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2002-11-19 | | Release date: | 2002-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Stepwise Adaptations of Citrate Synthase to Survival at Life'S Extremes. From Psychrophile to Hyperthermophile.
Eur.J.Biochem., 269, 2002
|
|
1SIB
 
 | |
1IF7
 
 | | Carbonic Anhydrase II Complexed With (R)-N-(3-Indol-1-yl-2-methyl-propyl)-4-sulfamoyl-benzamide | | Descriptor: | (R)-N-(3-INDOL-1-YL-2-METHYL-PROPYL)-4-SULFAMOYL-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IF9
 
 | | Carbonic Anhydrase II Complexed With N-[2-(1H-Indol-5-yl)-butyl]-4-sulfamoyl-benzamide | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-[2-(1H-INDOL-5-YL)-BUTYL]-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1I90
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH AL-8520 2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE, 4-AMINO-3,4-DIHYDRO-2-(3-METHOXYPROPYL)-, 1,1-DIOXIDE, (R) | | Descriptor: | 4-AMINO-6-[N-(3-METHOXYLPROPYL)-2H-THIENO[3,2-E][1,2]THIAZINE 1,1-DIOXIDE]-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chang, J.S, Liao, J, May, J.A, Christianson, D.W. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural aspects of isozyme selectivity in the binding of inhibitors to carbonic anhydrases II and IV.
J.Med.Chem., 45, 2002
|
|
1I9O
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,3,4-TRIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | 4-(AMINOSULFONYL)-N-[(2,3,4-TRIFLUOROPHENYL)METHYL]-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chandra, P.P, Jain, A, Christianson, D.W. | | Deposit date: | 2001-03-20 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fluoroaromatic-fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II.
J.Am.Chem.Soc., 123, 2001
|
|
1HVA
 
 | |
1HQX
 
 | | R308K ARGINASE VARIANT | | Descriptor: | ARGINASE, MANGANESE (II) ION | | Authors: | Lavulo, L.T, Sossong Jr, T.M, Brigham-Burke, M.R, Doyle, M.L, Cox, J.D, Christianson, D.W, Ash, D.E. | | Deposit date: | 2000-12-20 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Subunit-subunit interactions in trimeric arginase. Generation of active monomers by mutation of a single amino acid.
J.Biol.Chem., 276, 2001
|
|
8UT0
 
 | | Eukaryotic 80S ribosome with Reh1, eIF5A and A/P site tRNA | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Yelland, J.N, Taylor, D.W, Johnson, A.W. | | Deposit date: | 2023-10-30 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The Ribosome Assembly Factor Reh1 is Released from the Polypeptide Exit Tunnel in the Pioneer Round of Translation
To Be Published
|
|
8UTI
 
 | | Eukaryotic 80S ribosome with Reh1 and A/P site tRNA | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Yelland, J.N, Taylor, D.W, Johnson, A.W. | | Deposit date: | 2023-10-31 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | The Ribosome Assembly Factor Reh1 is Released from the Polypeptide Exit Tunnel in the Pioneer Round of Translation
To Be Published
|
|
1P8O
 
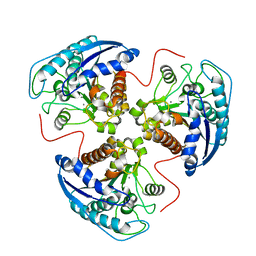 | | Structural and Functional Importance of First-Shell Metal Ligands in the Binuclear Manganese Cluster of Arginase I. | | Descriptor: | Arginase 1, MANGANESE (II) ION | | Authors: | Cama, E, Emig, F.A, Ash, D.E, Christianson, D.W. | | Deposit date: | 2003-05-07 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural and functional importance of first-shell metal ligands in the binuclear
manganese cluster of arginase I
Biochemistry, 42, 2003
|
|
1S8O
 
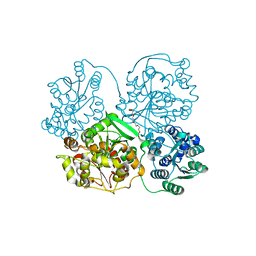 | | Human soluble Epoxide Hydrolase | | Descriptor: | HEXAETHYLENE GLYCOL, epoxide hydrolase 2, cytoplasmic | | Authors: | Gomez, G.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2004-02-03 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human epoxide hydrolase reveals mechanistic inferences on bifunctional catalysis in epoxide and phosphate ester hydrolysis
Biochemistry, 43, 2004
|
|
1A59
 
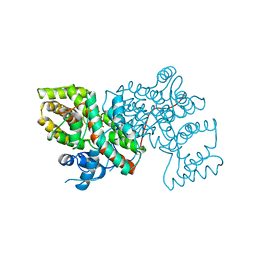 | | COLD-ACTIVE CITRATE SYNTHASE | | Descriptor: | CITRATE SYNTHASE, CITRIC ACID, COENZYME A | | Authors: | Russell, R.J.M, Gerike, U, Danson, M.J, Hough, D.W, Taylor, G.L. | | Deposit date: | 1998-02-20 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium.
Structure, 6, 1998
|
|
1HP0
 
 | | CRYSTAL STRUCTURE OF AN INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE FROM TRYPANOSOMA VIVAX IN COMPLEX WITH THE SUBSTRATE ANALOGUE 3-DEAZA-ADENOSINE | | Descriptor: | 3-DEAZA-ADENOSINE, CALCIUM ION, INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE | | Authors: | Versees, W, Decanniere, K, Pelle, R, Depoorter, J, Parkin, D.W, Steyaert, J. | | Deposit date: | 2000-12-12 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of a novel purine specific nucleoside hydrolase from Trypanosoma vivax.
J.Mol.Biol., 307, 2001
|
|
6JP4
 
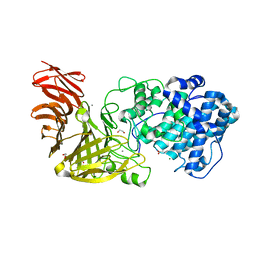 | | Crystal structure of the catalytic domain of a multi-domain alginate lyase Dp0100 from thermophilic bacterium Defluviitalea phaphyphila | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Alginate lyase, ... | | Authors: | Ji, S.Q, Dix, S.R, Aziz, A, Sedelnikova, S.E, Li, F.L, Rice, D.W. | | Deposit date: | 2019-03-25 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | The molecular basis of endolytic activity of a multidomain alginate lyase fromDefluviitalea phaphyphila, a representative of a new lyase family, PL39.
J.Biol.Chem., 294, 2019
|
|
5WDK
 
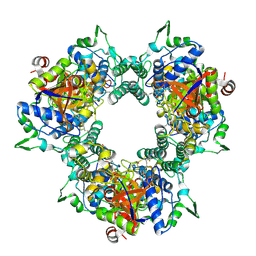 | | A processive dipeptidyl aminopeptidase secreted from an established commensal bacterium P. distasonis | | Descriptor: | 5-[(3aS,4R,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-N-(2-oxopropyl)pentanamide, Aminopeptidase C, POTASSIUM ION | | Authors: | Wolan, D.W, Xu, J.H, Solania, A, Chatterjee, S, Jiang, Z, ODonoghue, A.J. | | Deposit date: | 2017-07-05 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A Commensal Dipeptidyl Aminopeptidase with Specificity for N-Terminal Glycine Degrades Human-Produced Antimicrobial Peptides in Vitro.
Acs Chem.Biol., 13, 2018
|
|
1HQH
 
 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH NOR-N-HYDROXY-L-ARGININE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, NOR-N-OMEGA-HYDROXY-L-ARGININE | | Authors: | Cox, J.D, Cama, E, Colleluori, D.M, Ash, D.E, Christianson, D.W. | | Deposit date: | 2000-12-16 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase.
Biochemistry, 40, 2001
|
|
1O6T
 
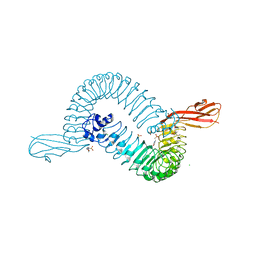 | | Internalin (INLA, Listeria monocytogenes) - functional domain, uncomplexed | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schubert, W.-D, Urbanke, C, Ziehm, T, Beier, V, Machner, M.P, Domann, E, Wehland, J, Chakraborty, T, Heinz, D.W. | | Deposit date: | 2002-10-15 | | Release date: | 2002-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Cell(Cambridge,Mass.), 111, 2002
|
|
1HMV
 
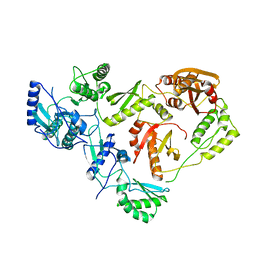 | | THE STRUCTURE OF UNLIGANDED REVERSE TRANSCRIPTASE FROM THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66), MAGNESIUM ION | | Authors: | Rodgers, D.W, Gamblin, S.J, Harris, B.A, Ray, S, Culp, J.S, Hellmig, B, Woolf, D.J, Debouck, C, Harrison, S.C. | | Deposit date: | 1994-12-15 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1R1O
 
 | |
1LV9
 
 | | CXCR3 Binding Chemokine IP-10/CXCL10 | | Descriptor: | Small inducible cytokine B10 | | Authors: | Booth, V, Keizer, D.W, Kamphuis, M.B, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2002-05-27 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The CXCR3 binding chemokine IP-10/CXCL10: structure and receptor interactions.
Biochemistry, 41, 2002
|
|
