3DXK
 
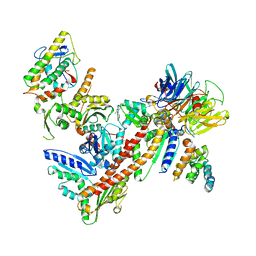 | | Structure of Bos Taurus Arp2/3 Complex with Bound Inhibitor CK0944636 | | Descriptor: | Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, Actin-related protein 2/3 complex subunit 2, ... | | Authors: | Nolen, B.J, Tomasevic, N, Russell, A, Pierce, D.W, Jia, Z, Hartman, J, Sakowicz, R, Pollard, T.D. | | Deposit date: | 2008-07-24 | | Release date: | 2009-07-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of two classes of small molecule inhibitors of Arp2/3 complex
Nature, 460, 2009
|
|
3H94
 
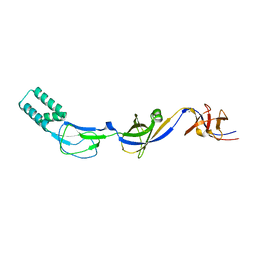 | | Crystal structure of the membrane fusion protein CusB from Escherichia coli | | Descriptor: | Cation efflux system protein cusB, SILVER ION | | Authors: | Su, C.-C, Yang, F, Long, F, Reyon, D, Routh, M.D, Kuo, D.W, Mokhtari, A.K, Van Ornam, J.D, Rabe, K.L, Hoy, J.A, Lee, Y.J, Rajashankar, K.R, Yu, E.W. | | Deposit date: | 2009-04-30 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Crystal structure of the membrane fusion protein CusB from Escherichia coli
J.Mol.Biol., 393, 2009
|
|
7UKK
 
 | |
6CW8
 
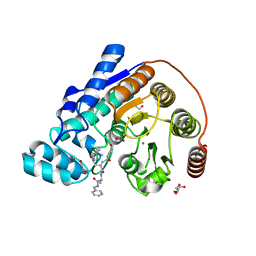 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with RTS-V5 | | Descriptor: | 1,2-ETHANEDIOL, Hdac6 protein, N-(2,2-dimethylpropyl)-N~2~-[4-(hydroxycarbamoyl)benzene-1-carbonyl]-L-asparaginyl-N-benzyl-L-alaninamide, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2018-03-30 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of the First-in-Class Dual Histone Deacetylase-Proteasome Inhibitor.
J. Med. Chem., 61, 2018
|
|
4GSM
 
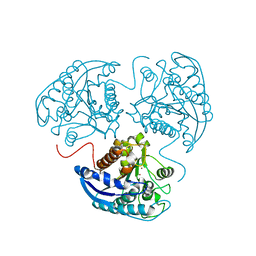 | |
4GWD
 
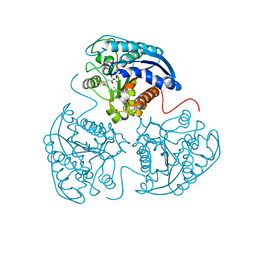 | | Crystal Structure of the Mn2+2,Zn2+-Human Arginase I-ABH Complex | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase-1, MANGANESE (II) ION, ... | | Authors: | D'Antonio, E.L, Hai, Y, Christianson, D.W. | | Deposit date: | 2012-09-01 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure and function of non-native metal clusters in human arginase I.
Biochemistry, 51, 2012
|
|
7UH4
 
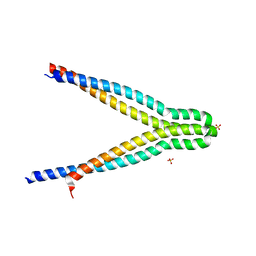 | | LXG-associated alpha-helical protein D2 (LapD2) | | Descriptor: | LXG-associated alpha-helical protein D2, SULFATE ION | | Authors: | Klein, T.A, Grebenc, D.W, Shah, P.Y, McArthur, O.D, Surette, M.G, Kim, Y, Whitney, J.C. | | Deposit date: | 2022-03-25 | | Release date: | 2022-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dual Targeting Factors Are Required for LXG Toxin Export by the Bacterial Type VIIb Secretion System.
Mbio, 13, 2022
|
|
7UJU
 
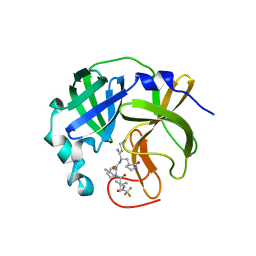 | | Room-temperature X-ray structure of monomeric SARS-CoV-2 main protease catalytic domain (MPro1-196) in complex with nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-03-31 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Autoprocessing and oxyanion loop reorganization upon GC373 and nirmatrelvir binding of monomeric SARS-CoV-2 main protease catalytic domain.
Commun Biol, 5, 2022
|
|
7UJ9
 
 | |
3HF5
 
 | | Crystal structure of 4-methylmuconolactone methylisomerase in complex with 3-methylmuconolactone | | Descriptor: | 4-methylmuconolactone methylisomerase, [(2S)-3-methyl-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Marin, M, Heinz, D.W, Pieper, D.H, Klink, B.U. | | Deposit date: | 2009-05-11 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and catalytic mechanism of 4-methylmuconolactone methylisomerase
J.Biol.Chem., 284, 2009
|
|
3HFK
 
 | | Crystal structure of 4-methylmuconolactone methylisomerase (H52A) in complex with 4-methylmuconolactone | | Descriptor: | 4-methylmuconolactone methylisomerase, [(2S)-2-methyl-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Marin, M, Heinz, D.W, Pieper, D.H, Klink, B.U. | | Deposit date: | 2009-05-12 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and catalytic mechanism of 4-methylmuconolactone methylisomerase
J.Biol.Chem., 284, 2009
|
|
7UJG
 
 | |
4HCE
 
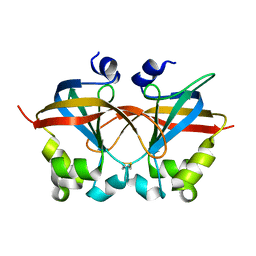 | | Crystal structure of the telomeric Saccharomyces cerevisiae Cdc13 OB2 domain | | Descriptor: | Cell division control protein 13 | | Authors: | Mason, M, Wannat, J.J, Harper, S, Schultz, D.C, Speicher, D.W, Johnson, F.B, Skordalakes, E. | | Deposit date: | 2012-09-29 | | Release date: | 2012-11-28 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cdc13 OB2 Dimerization Required for Productive Stn1 Binding and Efficient Telomere Maintenance.
Structure, 21, 2013
|
|
4GSZ
 
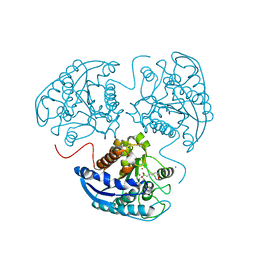 | |
4UTF
 
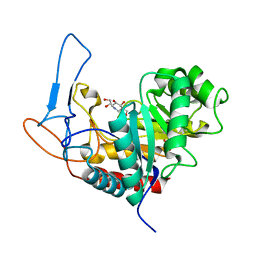 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-isofagomine and alpha- 1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, ... | | Authors: | Cuskin, F, Lowe, E.C, Temple, M.J, Zhu, Y, Pudlo, N.A, Cameron, E.A, Urs, K, Thompson, A.J, Cartmell, A, Rogowski, A, Tolbert, T, Piens, K, Bracke, D, Vervecken, W, Hakki, Z, Speciale, G, Munoz-Munoz, J.L, Pena, M.J, McLean, R, Suits, M.D, Boraston, A.B, Atherly, T, Ziemer, C.J, Williams, S.J, Davies, G.J, Abbott, D.W, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2014-07-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
6DKU
 
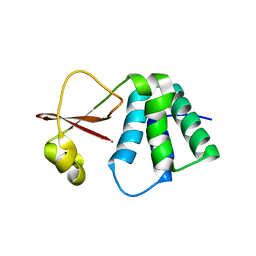 | | Crystal structure of Myotis VP35 mutant of interferon inhibitory domain | | Descriptor: | VP35 | | Authors: | Liu, H, Ginell, G.M, Keefe, L.J, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2018-05-30 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conservation of Structure and Immune Antagonist Functions of Filoviral VP35 Homologs Present in Microbat Genomes.
Cell Rep, 24, 2018
|
|
3CYU
 
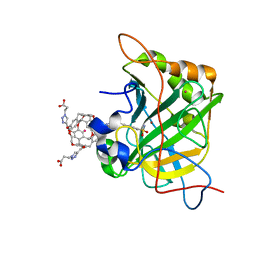 | | Human Carbonic Anhydrase II complexed with Cryptophane biosensor and xenon | | Descriptor: | Carbonic anhydrase 2, MoMo-2-[4-(2-(4-(methoxy)-1H-1,2,3-triazol-1-yl)ethyl)benzenesulfonamide]-7,12-bis-[3-(4-(methoxy)-1H-1,2,3-triazol-1-y l)propanoic acid]-cryptophane-A, PoPo-2-[4-(2-(4-(methoxy)-1H-1,2,3-triazol-1-yl)ethyl)benzenesulfonamide]-7,12-bis-[3-(4-(methoxy)-1H-1,2,3-triazol-1-yl)propanoic acid]-cryptophane-A, ... | | Authors: | Aaron, J.A, Jude, K.M, Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2008-04-26 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a 129Xe-cryptophane biosensor complexed with human carbonic anhydrase II.
J.Am.Chem.Soc., 130, 2008
|
|
6DVM
 
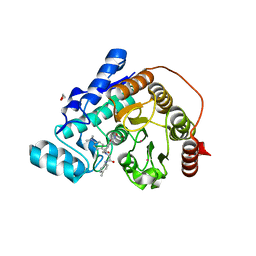 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with DDK-122 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(dimethylamino)-N-{[4-(hydroxycarbamoyl)phenyl]methyl}-N-{2-[(4-methylphenyl)amino]-2-oxoethyl}benzamide, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Histone Deacetylase 6-Selective Inhibitors and the Influence of Capping Groups on Hydroxamate-Zinc Denticity.
J. Med. Chem., 61, 2018
|
|
6DVO
 
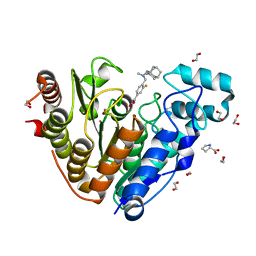 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with Bavarostat | | Descriptor: | 1,2-ETHANEDIOL, 3-fluoro-N-hydroxy-4-[(methyl{[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl]methyl}amino)methyl]benzamide, Hdac6 protein, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Histone Deacetylase 6-Selective Inhibitors and the Influence of Capping Groups on Hydroxamate-Zinc Denticity.
J. Med. Chem., 61, 2018
|
|
4EPH
 
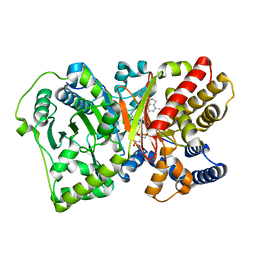 | | CRYSTAL STRUCTURE OF RAT CARNITINE PALMITOYLTRANSFERASE 2 IN COMPLEX with CoA-site inhibitor | | Descriptor: | 2-chloro-4-[({1-[(5-chloro-2-methoxyphenyl)sulfonyl]-4-methyl-2,3-dihydro-1H-indol-6-yl}carbonyl)amino]benzoic acid, Carnitine O-palmitoyltransferase 2, mitochondrial, ... | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2012-04-17 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Isothermal titration calorimetry with micelles: Thermodynamics of inhibitor binding to carnitine palmitoyltransferase 2 membrane protein.
FEBS Open Bio, 3, 2013
|
|
7TPS
 
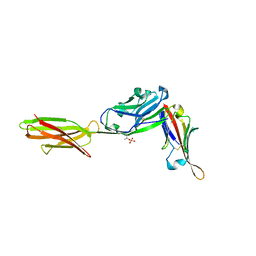 | | Crystal structure of ALPN-202 (engineered CD80 vIgD) in complex with PD-L1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Demonte, D.W, Maurer, M.F, Akutsu, M, Kimbung, Y.R, Logan, D.T, Walse, B. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The engineered CD80 variant fusion therapeutic davoceticept combines checkpoint antagonism with conditional CD28 costimulation for anti-tumor immunity.
Nat Commun, 13, 2022
|
|
7U69
 
 | |
7U3M
 
 | |
7U6A
 
 | |
3E9B
 
 | |
