4QA6
 
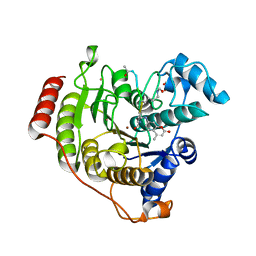 | | Crystal structure of I243N/Y306F HDAC8 in complex with a tetrapeptide substrate | | Descriptor: | 7-AMINO-4-METHYL-CHROMEN-2-ONE, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
4MU4
 
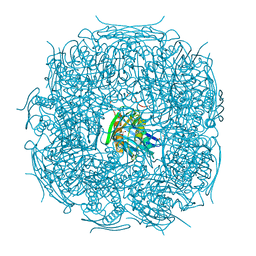 | | The form B structure of an E21Q catalytic mutant of A. thaliana IGPD2 in complex with Mn2+ and its substrate, 2R3S-IGP, to 1.41 A resolution | | Descriptor: | (2R,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate, 1,2-ETHANEDIOL, Imidazoleglycerol-phosphate dehydratase 2, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
4MYL
 
 | |
4Q3Q
 
 | | Crystal structure of Schistosoma mansoni arginase in complex with inhibitor ABH | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase, GLYCEROL, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
4QA2
 
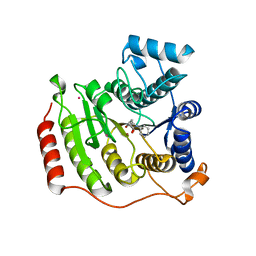 | | Crystal structure of I243N HDAC8 in complex with SAHA | | Descriptor: | GLYCEROL, Histone deacetylase 8, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.377 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
4Q3S
 
 | | Crystal structure of Schistosoma mansoni arginase in complex with inhibitor ABHPE | | Descriptor: | Arginase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
4Q3T
 
 | | Crystal structure of Schistosoma mansoni arginase in complex with inhibitor NOHA | | Descriptor: | Arginase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
4QA1
 
 | | Crystal structure of A188T HDAC8 in complex with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
1CVF
 
 | |
1CWU
 
 | | BRASSICA NAPUS ENOYL ACP REDUCTASE A138G MUTANT COMPLEXED WITH NAD+ AND THIENODIAZABORINE | | Descriptor: | 6-METHYL-2(PROPANE-1-SULFONYL)-2H-THIENO[3,2-D][1,2,3]DIAZABORININ-1-OL, ENOYL ACP REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Roujeinikova, A, Rafferty, J.B, Rice, D.W. | | Deposit date: | 1999-08-26 | | Release date: | 1999-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitor binding studies on enoyl reductase reveal conformational changes related to substrate recognition.
J.Biol.Chem., 274, 1999
|
|
6U4Z
 
 | |
1CVA
 
 | |
1CNX
 
 | |
5BWZ
 
 | |
6UFN
 
 | |
6UHV
 
 | |
6UII
 
 | |
1CR6
 
 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE COMPLEXED WITH CPU INHIBITOR | | Descriptor: | EPOXIDE HYDROLASE, N-CYCLOHEXYL-N'-(PROPYL)PHENYL UREA | | Authors: | Argiriadi, M.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 1999-08-13 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CVC
 
 | |
1CIN
 
 | | THE POSITIONS OF HIS-64 AND A BOUND WATER IN HUMAN CARBONIC ANHYDRASE II UPON BINDING THREE STRUCTURALLY RELATED INHIBITORS | | Descriptor: | (4S-TRANS)-4-(METHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, CARBONIC ANHYDRASE II, METHYL MERCURY ION, ... | | Authors: | Smith, G.M, Alexander, R.S, Christianson, D.W, Mckeever, B.M, Ponticello, G.S, Springer, J.P, Randall, W.C, Baldwin, J.J, Habecker, C.N. | | Deposit date: | 1993-10-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Positions of His-64 and a bound water in human carbonic anhydrase II upon binding three structurally related inhibitors.
Protein Sci., 3, 1994
|
|
1CNY
 
 | |
1CNG
 
 | |
1CUK
 
 | |
1CNK
 
 | |
1CIL
 
 | | THE POSITIONS OF HIS-64 AND A BOUND WATER IN HUMAN CARBONIC ANHYDRASE II UPON BINDING THREE STRUCTURALLY RELATED INHIBITORS | | Descriptor: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Smith, G.M, Alexander, R.S, Christianson, D.W, Mckeever, B.M, Ponticello, G.S, Springer, J.P, Randall, W.C, Baldwin, J.J, Habecker, C.N. | | Deposit date: | 1993-10-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Positions of His-64 and a bound water in human carbonic anhydrase II upon binding three structurally related inhibitors.
Protein Sci., 3, 1994
|
|
